
Understanding the mechanism of early tumor occurrence is crucial to achieving early prevention and treatment of cancer. The traditional Peter Nowell clonal evolution theory holds that tumors are caused by a single cell with oncogenic mutations, and after continuous cell expansion and screening, malignant tumors are finally formed. This theory emphasizes that tumors have a "monoclonal origin", that is, the tumor originates from a single cell and expands. However, our understanding of the precancerous process of tumors at earlier stages is still very limited. For example, in a precancerous lesion, are there multiple populations of cells with independent mutations that work together to drive tumor development (i.e., "polyclonal origin")? There is no clear answer to this question at this time. In general, the lack of understanding of the origin and mechanism of early tumors has greatly limited our breakthroughs in precise tumor screening and early intervention.
At 24:00 on October 30, Beijing time, Hu Zheng from the Institute of Synthetic Biology of Shenzhen Institute of Advanced Technology, Chinese Academy of Sciences, He Xionglei and He Zhen from Sun Yat-sen University published a research paper entitled "Polyclonal-to-monoclonal transition in colorectal precancerous evolution" online in Nature, revealing for the first time a new early evolutionary model of tumor transition from polyclonal to monoclonal. The mechanism of cell-to-cell communication and interaction in the origin of tumor polyclonal origin was also analyzed. This discovery not only provides a new scientific perspective for the early pathogenesis mechanism of tumors, but also puts forward new ideas for accurate early screening and intervention of cancer.

Figure 1. Screenshot of the article online
For the first time in mammals, the research team has established a base-editor-based cell lineage tracing technology (called SMALT) to achieve high-precision lineage tracking with single-cell resolution in mice. SMALT technology combines cytidine deaminase (AID), artificial DNA barcoding sequences, and the Tet-On inducible expression system to enable large-scale, highly accurate reconstruction of single-cell lineage trees through AID-mediated targeted mutations and subsequent DNA sequencing (Figure 2a). Using transgenic mice carrying the SMALT system, the research team constructed two classical mouse bowel cancer models: an inflammation-induced bowel cancer model (AOM/DSS model) and a multiple polyp model (ApcMin/+ model). In the lineage tree analysis, the team found that the majority (66.7%) of inflammation-induced bowel cancers and all Apc intestinal polyps exhibited polyclonal origin (Fig. 2b). By integrating DNA barcoding mutations, whole-genome sequencing, and single-cell transcriptome data, the research team further found that monoclonal tumors (Fig. 2c) have a higher degree of malignancy than polyclonal tumors, suggesting that monoclonal tumors may represent a more "advanced" stage of tumor progression. Based on these findings, the team proposed a new model of "polyclonal to monoclonal transition" of tumorigenesis (Fig. 2D), which provides a new theoretical framework for early tumor evolution.
The research team also collected a cohort of 107 human patients with sporadic bowel polyps and colorectal cancer. Genomic sequencing data show that approximately 30% of human intestinal polyps are of polyclonal origin and have low mutational load and copy number variation. Pathological features also indicate that polyclonal polyps are smaller in size and less malignant than monoclonal polyps. In contrast, monoclonal polyps exhibit more genomic variants, larger sizes, and higher levels of malignancy. These findings validate lineage tracing results in mice in humans, and together support an early tumor evolutionary pattern of polyclonal to monoclonal transition.
Based on these results, the research team proposed a new model of polyclonal-monoclonal transition in early tumorigenesis (Fig. 2D), emphasizing the importance of specific genomic and microenvironmental changes and cell-cell interactions in this process. These findings provide a new conceptual framework for understanding the origin of tumors and propose new strategies for tumor prevention by targeting cell-to-cell communication to achieve early intervention. Therefore, this study opens up a new field of research, that is, to explore the mechanism of polyclonal formation and cell-to-cell interactions in early tumorigenesis, and provides a new idea for early tumor screening, risk prediction and early targeted intervention.
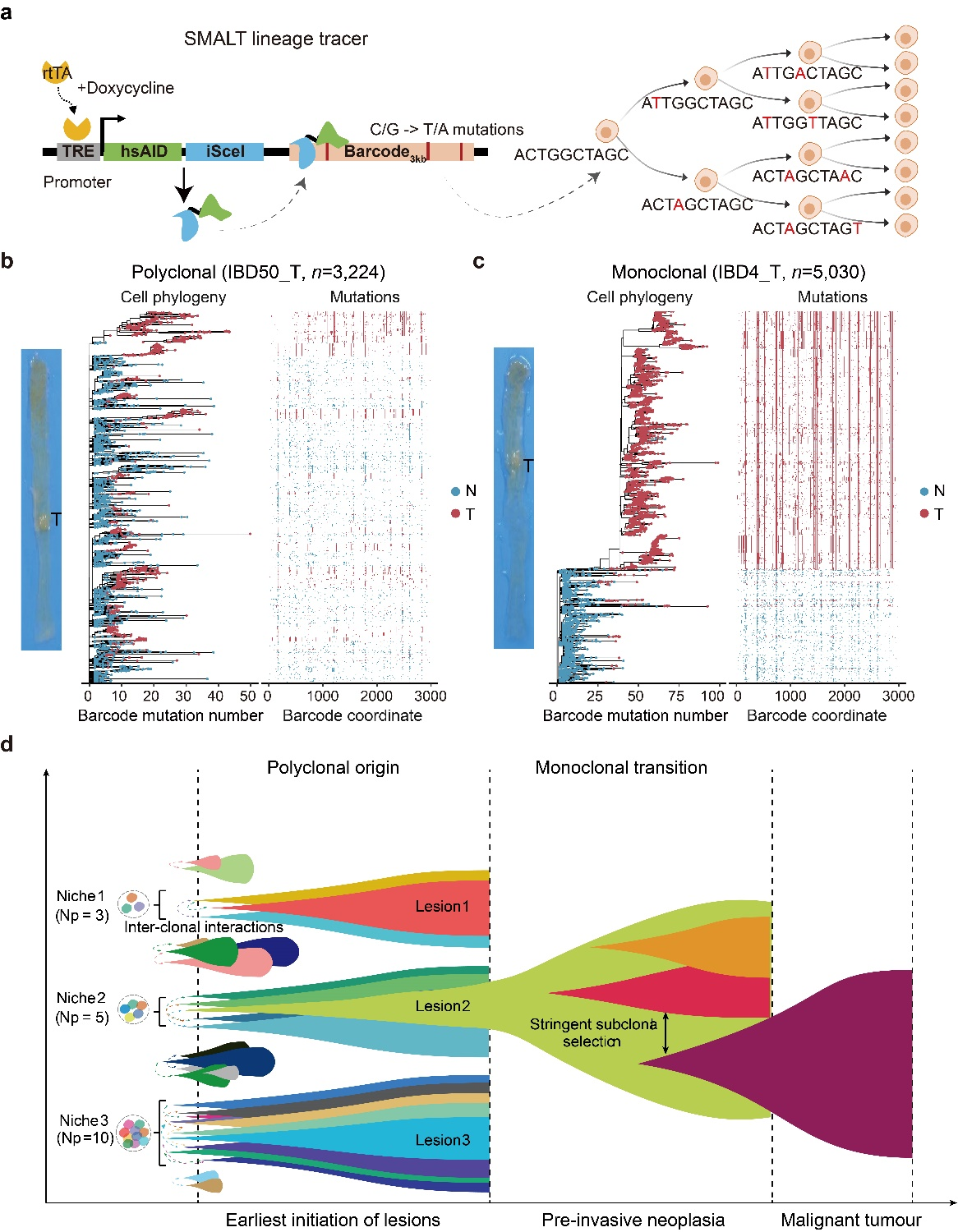
Figure 2. Single-cell lineage traces a new evolutionary pattern of early tumor transition from polyclonal to monoclonal.
Hu Zheng, a researcher at the Institute of Synthetic Biology, Shenzhen Institute of Advanced Technology, Chinese Academy of Sciences, Professor He Xionglei from the School of Life Sciences, Sun Yat-sen University, and Professor He Zhen from the Sixth Affiliated Hospital of Sun Yat-sen University are the co-corresponding authors of this paper. Dr. Lu Zhaolian and Dr. Mo Shanlan, assistant researchers of Shenzhen Institute of Advanced Technology, Chinese Academy of Sciences; Xie Duo, a doctoral student jointly trained by Shenzhen Institute of Advanced Technology, Chinese Academy of Sciences and University of Macau; Dr. Zhai Xiangwei, a doctoral student at Sun Yat-sen University, and Dr. Deng Shanjun, a postdoctoral fellow, are the co-first authors of the paper. The research results were supported by the National Key R&D Program of China, the National Natural Science Foundation of China, the Chinese Academy of Sciences, the Natural Science Foundation of Guangdong Province, and the Shenzhen Institute of Synthetic Biology.
Original link: https://www.nature.com/articles/s41586-024-08133-1
Schematic diagram of the paper:

Caption: The color of peacock feathers gradually changed from colorful to monoclonal, expressing the process of tumor cell evolution from polyclonal to monoclonal state.
Expert commentary
Qimin Zhan (Academician of the Chinese Academy of Engineering, Boya Chair Professor, Peking University, Molecular Oncologist)
The study published in Nature by the team of Zheng Hu, Xionglei He and Zhen He marks a new level in the understanding of the early evolution of tumors. This study is the first to propose an early tumor evolution model from polyclonal to monoclonal transition, and reveals the important mechanism of cell-to-cell communication and microenvironment changes in this process. This discovery not only breaks through the classical monoclonal origin theory, but also expands our understanding of the origin and evolution of tumor heterogeneity, and opens up a new direction for the early intervention strategy of precision medicine.
The phenomenon of polyclonal origin of tumors was systematically confirmed for the first time in this study. Using high-resolution cell lineage tracing technology and single-cell transcriptome sequencing, the research team observed in mouse models and human tissues of precancerous lesions that early-stage tumor lesions often have multiple independent sources of cell clones, which work together to promote disease progression through cell-to-cell communication and cooperation in the early stages of tumorigenesis. As tumors progress, these polyclonals are gradually replaced by a dominant clone and transformed into monoclonal tumors, a dynamic process that is particularly evident in genomic level and microenvironment interactions. The analysis of this mechanism provides a new theoretical basis for the staging, prediction and individualized treatment strategies of tumors.
From the perspective of precision medicine, this discovery is of great significance for the early diagnosis and intervention of tumors. Traditional early-stage tumor screening has relied primarily on the detection of markers of a single driver mutation, however this approach may have limitations in tumors of polyclonal origin. The mechanism of polyclonal origin and cell-to-cell communication revealed in this study suggests that we need to pay attention to multiple gene mutations and cell-to-cell interaction networks to identify polyclonal lesions with the potential for progression. This provides an important basis for the development of interventions that target the polyclonal stage, such as strategies to stop tumor evolution by inhibiting cell-to-cell communication pathways.
All in all, this study provides a new idea for early screening, prevention and individualized intervention of tumors. Through an in-depth understanding of the genomic characteristics of early tumor evolution, we are expected to achieve more accurate early precancerous lesion detection and risk prediction, promote the advancement of tumor precision medicine, and provide a scientific basis for cancer prevention and treatment.
Wu Zhongyi (Chair Professor of Sun Yat-sen University, Academician of Academia Sinica, Taiwan, Former Dean of the Department of Evolutionary Ecology, University of Chicago, Former Director of the Beijing Institute of Genomics, Chinese Academy of Sciences)
In the 70s of the 20th century, Peter Nowell first systematically proposed the clonal evolution theory of tumors, which believed that tumors are cell populations of single origin but developed a high degree of genetic diversity, which is a clonal evolution process driven by Darwin's natural selection. This theory was proposed at a time of rapid development in molecular biology and genetics, and the rapid rise of technologies such as DNA sequencing has greatly promoted people's understanding of tumor biology, clonal heterogeneity and evolutionary mechanisms.
However, the hypothesis of monoclonal or polyclonal origin in the early stages of tumor evolution has been debated. On the one hand, genome sequencing appears to support Nowell's view of a monoclonal origin. Sequencing data, on the other hand, only show that the vast majority of cancer cells belong to the same clone. Under cell competition, other clones are difficult to detect after the tumor begins to grow. At the same time, the theory of field cancerization also puts forward the opposite view, that is, a microenvironment that can produce cell clones should be able to produce more than one cell clone. In 2022, Sun Yat-sen University Bingjie Chen et al. Natl Sci Rev 2022 also supported this view of polyclonal origin.
In recent years, the development and application of new technologies such as lineage tracing technology have brought the study of tumor evolution to a new stage. In this paper, the joint team of Hu Zheng, He Xionglei and He Zhen skillfully used the technology of lineage tracing to observe the multiple origins of tumor clones from the source. They found that a polyclonal pattern of origin was prevalent in early bowel cancer in mice and humans. Although polyclonal origins have occasionally been found in past studies, Hu et al. concluded that polyclonal origins are very common. Essentially a significant extension of the single origin doctrine of Peter Nowell's clonal evolutionary theory.
Another conclusion of Hu Zheng et al. is that cells in polyclonal tumors seem to cooperate with each other rather than compete with each other. This argument can be thought of from many angles. Cooperation in the evolutionary process is generally limited to the same genotype. It is unclear whether their observations conform to this routine. The driving mechanism behind this requires further research. In conclusion, the proposal of this new theoretical paradigm will not only promote the progress of somatic cell evolution, but also provide new guidance for the early diagnosis and personalized treatment of tumors.
Bai Fan (Professor, Peking University, Xplorer Prize Winner)
The traditional theory of tumor clonal evolution holds that tumors are initiated by a single mutant cell and eventually formed through continuous clonal expansion and selection. The Nature paper by Hu Zheng from the Shenzhen Institute of Advanced Technology, Chinese Academy of Sciences, He Xionglei and He Zhen from Sun Yat-sen University revealed for the first time the phenomenon of polyclonal origin in the precancerous lesion stage, and proposed a new model of early tumor transition from polyclonal to monoclonal, which provides an important scientific breakthrough for understanding the polyclonal to monoclonal transition process that occurs in the early stage of tumors, and is of great significance for in-depth understanding of the early dynamic evolution of tumors.
The research team used a combination of high-resolution cell lineage tracing, single-cell transcriptome sequencing, and genomic sequencing of population samples in mouse models to provide a multi-dimensional insight into the mechanisms of cell communication and interaction in early polyclonal lesions. It has been found that cell-to-cell communication decreases in the progression of early polyclonal lesions to monoclonality, and this cell-to-cell interaction may be an important driving force in polyclonal formation. This discovery not only provides a new conceptual framework for tumor origin, but also points to the key role of microenvironment and genomic alterations in tumor evolution.
The scientific significance of this study lies in the fact that it provides a new strategy for early tumor screening and targeted prevention. By targeting the cellular communication network in early precancerous lesions, it is expected that effective intervention can be carried out before the tumor enters the monoclonal stage, thereby preventing the further malignant evolution of the tumor. This research not only promotes the cutting-edge development of basic oncology, but also provides practical guidance for precision medicine and cancer prevention, which may have an important impact on future early tumor screening technology and risk prediction. At the same time, this study comprehensively uses a variety of cutting-edge technologies, including single-cell lineage tracing and animal models, population cohort studies, single-cell transcriptomes, intestinal single crypt sequencing, organoids, quantitative modeling, etc., representing a new paradigm of multidisciplinary research in oncology.