
On October 10th Beijing time, a collaborative effort between Dr. Lou Chunbo's research group at the Shenzhen Institutes of Advanced Technology, Chinese Academy of Sciences, and Dr. Wu Qiong's research group at the School of Life Sciences, Tsinghua University, resulted in a groundbreaking study published in Nature Communications. The research presents a new genetic circuit capable of dynamically sensing and activating any RNA expression, titled "High-resolution and programmable RNA-IN and RNA-OUT genetic circuit in living mammalian cells." This work achieves the reconstruction of any endogenous RNA regulatory network within living mammalian cells by constructing an RNA-IN/RNA-OUT genetic circuit with sensing and response functions. It also has the ability to sense point mutations in RNA sequences within living cells, marking the first time that single-point mutation sensing capability has been enhanced from 1.5 times to 94 times. This genetic circuit demonstrates broad application potential in various cell therapy and gene therapy scenarios, including stem cell differentiation state sensing, intracellular activation of endogenous progesterone synthetic metabolic pathways, and the identification and selective killing of tumor cells with point mutations, providing a new toolkit for precise control of cell fate.
Dr. Zhang Min and Dr. Zhang Xue from Tsinghua University are the co-first authors, and Dr. Lou Chunbo from the Shenzhen Institutes of Advanced Technology, Chinese Academy of Sciences, and Associate Professor Wu Qiong from Tsinghua University are the co-corresponding authors of the paper. Ph.D. student Xu Yongyue and Zhang Bo from Tsinghua University, and Dr. Xiang Yanhui from the Shenzhen Institutes of Advanced Technology, Chinese Academy of Sciences, made significant contributions.

Article Online Screenshot
Original Article Link: https://doi.org/10.1038/s41467-024-52962-7
The rapid development of single-cell technologies and various cell atlas projects has provided a wealth of molecular tags for deciphering cell types and states across multiple spatial and temporal dimensions. RNA, as the core medium determining the diversity of cell types and states, can lead to the evolution of various pathological cell states, including tumorigenesis, when its expression is dysregulated or mutated. However, current technologies are mostly limited to RNA detection or tracing and cannot directly convert RNA change signals into regulatory signals for cell state control. Although the latest RNA sensors, such as eToehold switch and ADAR switch, have filled the gap in the field of RNA concentration sensing, they are not flexible enough to sense point mutations and have limitations such as poor designability or off-target toxicity. How to precisely, efficiently, and programmatically sense the dynamic changes of a wide spectrum of RNA concentrations and sequences in living cells and manipulate specific target cells remains a key challenge in the fields of life sciences and medicine.
This study proposes a new strategy to develop a highly sensitive, programmable, and single-nucleotide resolution RNA sensing and response genetic circuit within living cells, named RNA-IN/RNA-OUT. The circuit mainly includes three modules: an upstream RNA recognition sensing module responsible for endogenous RNA input (achieving RNA-IN), a downstream effector module responsible for endogenous RNA expression output (achieving RNA-OUT), and a processing module responsible for presenting RNA information (Figure 1).
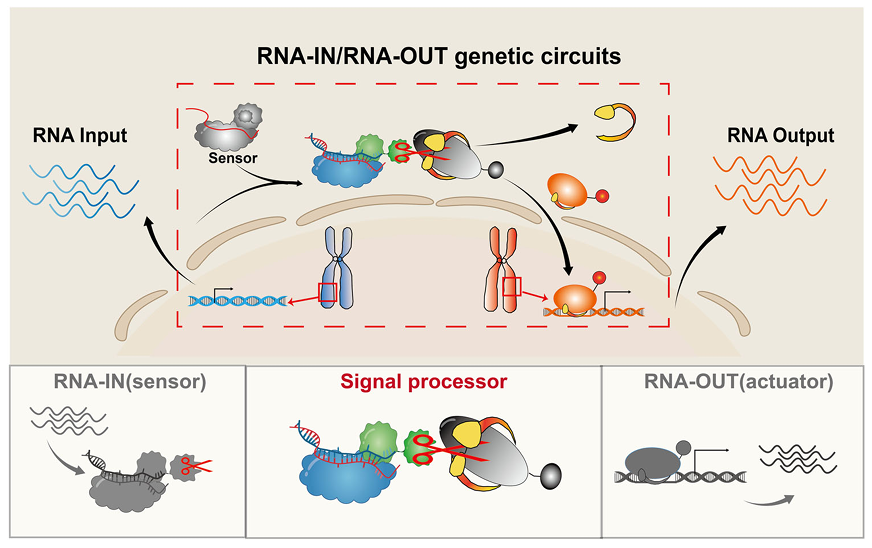
Figure 1. Schematic diagram of the working principle of RNA-IN/RNA-OUT
Researchers first constructed a programmable RNA sensor named CASP sensor, which plays the role of sensing dynamic RNA signals and forms the RNA-IN module. The CASP sensor consists of two parts: a programmable RNA-binding protein (DiCas7-11) and an effector protein (transcription activator CI434 and activation domain VP64). The CASP sensor follows the crRNA to guide the activation of the protease activity, releasing the effector protein anchored on the cell membrane to achieve transcriptional regulation. On the basis of precise design, in order to achieve sensitive sensing of a wide spectrum of RNAs, the research team systematically adjusted and optimized each component in the CASP sensor, successfully detecting endogenous expression as low as 8 transcripts per million transcripts (TPM) (Figure 2).
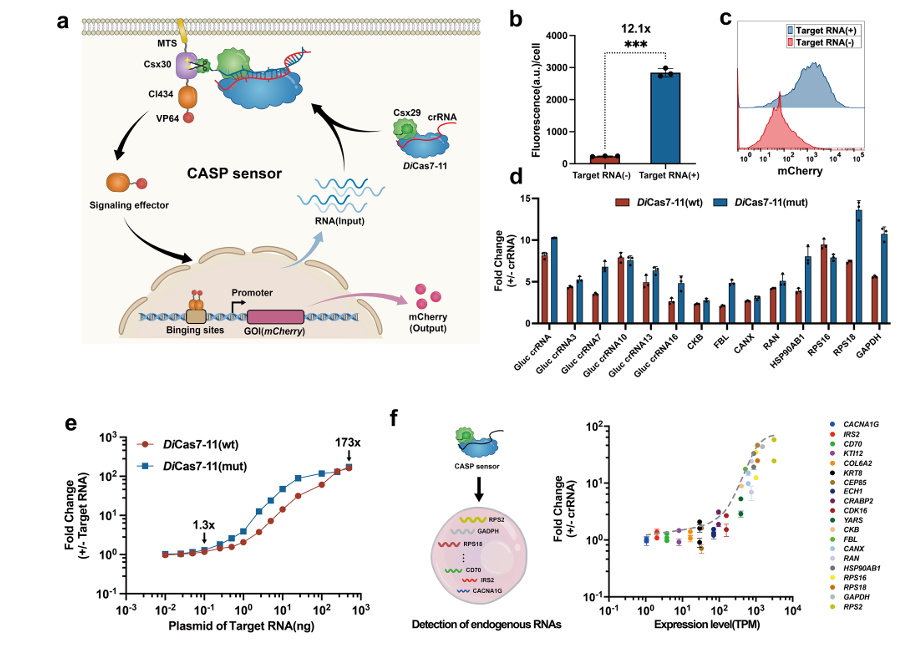
Figure 2. Design and characterization of programmable CASP sensors (RNA-IN)
In addition to RNA expression abnormalities, gene mutations are also an important cause of somatic diseases such as cancer, vascular, and neurological diseases. However, there is often only a small difference of a single nucleotide mutation between the wild-type and mutant RNA sequences, which is often difficult to detect and poses a more challenging requirement for the sensing resolution of the RNA-IN module. However, the key element of RNA sensing in the CASP sensor, "DiCas7-11," has a high tolerance for single base mismatches and is not sufficient for the detection of single-point mutations. The research team systematically explored the critical point of DiCas7-11's tolerance to base mismatches, and by introducing a synergistic strategy with auxiliary mutation sites, a single base mismatch difference was formed between the crRNA and the target RNA sequence, enhancing the originally undetectable single-point mutation RNA sensing from 1.5 times to 94 times. Thus, the detection of single-base mutations was successfully achieved, and the sensing of RNA expression was expanded to the sensing of sequence changes, greatly enriching the recognition range of the RNA-IN module. In particular, in the detection of representative point mutations in tumor key genes such as KRAS, TP53, BRAF, PIK3CA, and EGFR, the CASP sensor demonstrated sensitive recognition capabilities (Figure 3).
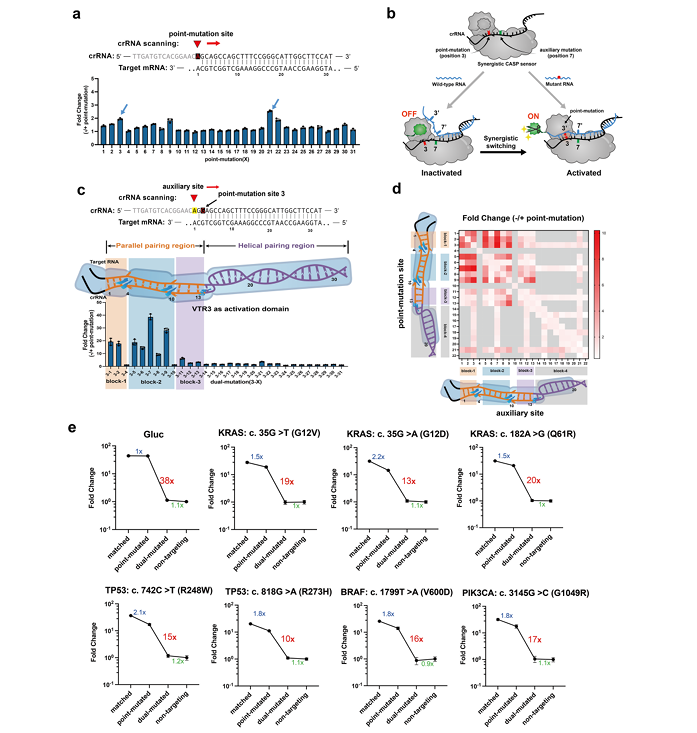
Figure 3. Synergistic CASP sensor sensing and response to single-nucleotide mutations (RNA-IN)
Furthermore, the research team connected the CASP sensor (RNA-IN) with the programmable dSpCas9-VPR endogenous activator (RNA-OUT) to form the complete RNA-IN/RNA-OUT genetic circuit. Using this genetic circuit to sense different expression levels of RNA within cells, as well as the dynamic changes of RNA within cells stimulated by environmental factors, the team ultimately achieved the transcriptional activation of specific genes (Figure 4).
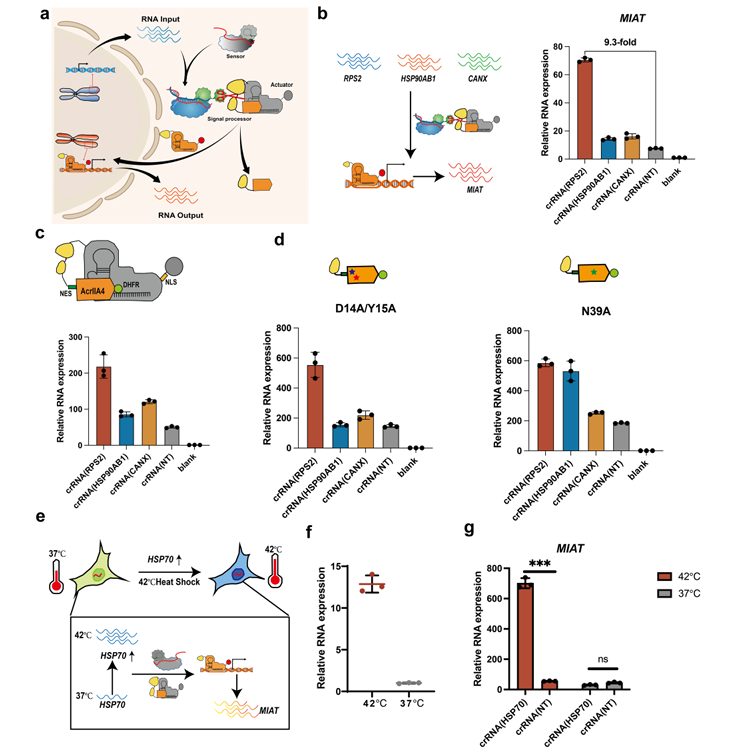
Figure 4. Design and optimization of the RNA-IN/RNA-OUT genetic circuit
Finally, researchers fully demonstrated the ultra-sensitive perception and flexible manipulation capabilities of the RNA-IN/RNA-OUT genetic circuit for endogenous RNA in different cell types, including: (1) connecting continuously expressed RNA to activate the endogenous biosynthetic metabolic network of progesterone; (2) dynamically monitoring cell state changes during cell differentiation and transdifferentiation; (3) identifying characteristic point mutations in RNA to selectively kill pancreatic and liver cancer cells (Figure 5).
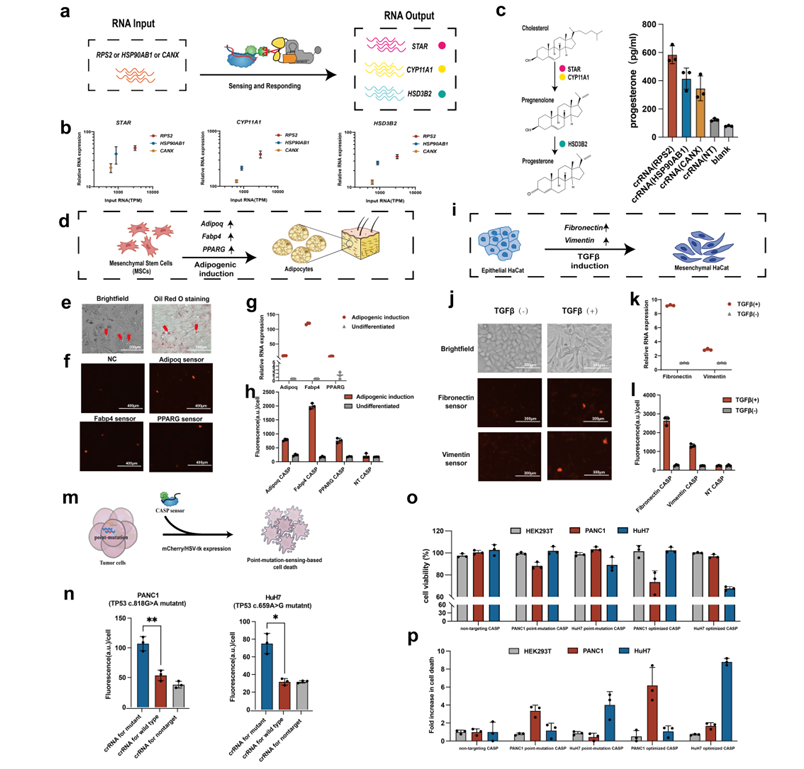
Figure 5. Application expansion of the RNA-IN/RNA-OUT genetic circuit
In summary, the RNA-IN/RNA-OUT genetic circuit features high sensitivity, programmability, and single-nucleotide resolution. This circuit senses RNA dynamics within living cells and directly converts them into transcriptional regulatory commands for specific genes, establishing a strong correlation between any RNAs, with the potential to reconstruct the internal RNA regulatory network of cells and endow cells with new biological functions. The circuit has a broad application prospect in the fields of cell and gene therapy, cellular reprogramming, and the biosynthesis of compounds, providing revolutionary technical support for the manipulation of cell fate.
The study was supported by the Ministry of Science and Technology of China, the National Natural Science Foundation, the Chinese Academy of Sciences, and the Vanke Public Health and Health Discipline Development Special Fund of Tsinghua University.