
Patients receiving organ transplants are in a long-term immunocompromised state and are prone to various severe infections, including invasive fungal infections. Therefore, these patients need to continuously use antibacterial and antifungal drugs to prevent infections. However, even with drug protection, breakthrough infections during medication are still frequently observed in clinical settings, and the microorganisms causing these infections do not show classical antimicrobial resistance to the used drugs. Due to the unclear mechanism of these infections, it is difficult to quickly find effective treatment plans, leading to a high mortality rate among patients. Therefore, there is an urgent need to analyze the etiology through high-quality clinical data and experimental data, develop new detection methods, and address this challenging medical problem.
On August 2, 2024, the team led by Zhai Bing at the Institute of Synthetic Biology of the SIAT, in collaboration with Dr. Liao Chen from Memorial Sloan Kettering Cancer Center, Tobias Hohl's team, and David Weiss's team from Emory University, published their latest research findings titled "Antifungal heteroresistance causes prophylaxis failure and facilitates breakthrough Candida parapsilosis infections" in Nature Medicine.

Screenshot of the article online
Link:https://www.nature.com/articles/s41591-024-03183-4
The study found that many strains of Candida parapsilosis, which cause breakthrough fungemia in patients receiving antifungal prophylaxis with micafungin, exhibit a phenotype distinct from classical resistance known as heteroresistance. Patients colonized with heteroresistant strains have a significantly higher risk of developing breakthrough infections compared to those colonized with only susceptible strains. The research team, in collaboration with the China Hospital Invasive Fungi Surveillance Net (CHIF-NET) and several groups in North America and Europe, discovered that this clinically significant drug sensitivity phenotype is prevalent among strains worldwide. Existing clinical susceptibility testing methods misidentify heteroresistant strains as sensitive, leading to incorrect selection of antifungal drugs. Using machine learning models, the study accurately predicted the heteroresistance phenotype based on no more than 10 genomic features. This research highlights the clinical importance of heteroresistance and provides new strategies for developing effective rapid drug sensitivity tests.
Antimicrobial resistance causes over a million deaths globally each year, posing a severe public health challenge. Heteroresistance differs from classical resistance phenotypes by describing the coexistence within the same strain of a minority of resistant cells (sometimes as low as one in a million or even less) alongside the majority of sensitive cells. At higher drug concentrations, most sensitive cells are killed while resistant cells survive and proliferate. Due to the low frequency of resistant cells, heteroresistance cannot be identified by conventional clinical susceptibility testing methods. In critical infections, multiple antibiotics are often used simultaneously, making it difficult to directly link clinical outcomes to the drug sensitivity phenotype of a single agent and thus clarify the real-world significance of heteroresistance. In contrast, the preventive use of a single antimicrobial agent in immunocompromised patients offers a better opportunity to study the clinical relevance of drug sensitivity phenotypes.
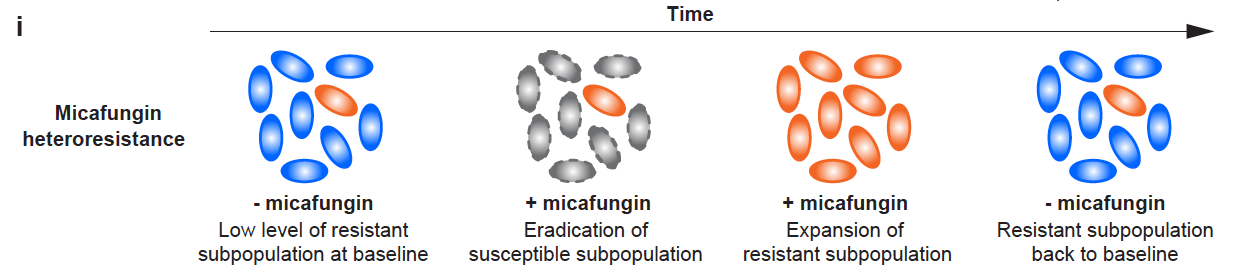
Figure 1. The proportion of drug-resistant cells in heteroresistant strains varies with drug exposure conditions
The authors first conducted a retrospective analysis of breakthrough fungemia cases that occurred among 952 patients who underwent hematopoietic stem cell transplantation at Memorial Sloan Kettering Cancer Center (MSKCC) between 2016 and 2020, finding that nearly half were caused by Candida parapsilosis. These patients had all received micafungin (a member of the echinocandin class of drugs targeting fungal cell wall synthesis) for fungal infection prophylaxis during their transplant period. Notably, the C. parapsilosis strains causing these breakthrough infections were identified as micafungin-sensitive. Therefore, the researchers performed more precise population analysis profiling (PAP) on these strains and discovered that most exhibited heteroresistance to micafungin.
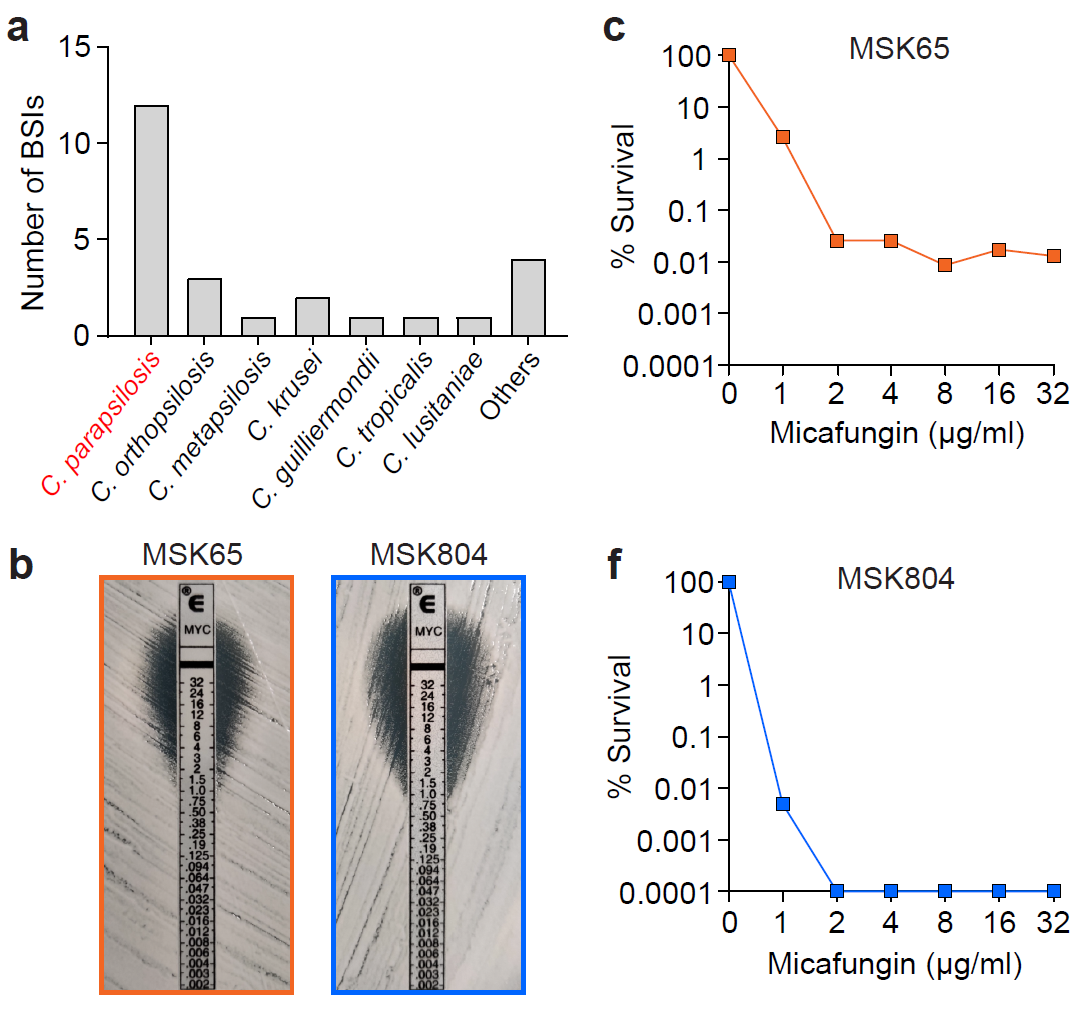
Figure 2. Breakthrough fungemia cases in MSKCC are predominantly caused by heterogeneous drug-resistant strains (orange)
In previous studies, the team led by Zhai Bing identified that colonization of Candida parapsilosis in the gut is a necessary condition for the development of fungemia based on the same clinical cohort (Nature Medicine 2020), but only a subset of patients with gastrointestinal colonization of C. parapsilosis during transplantation developed fungal infections (Nature Microbiology 2021). The authors of this study conducted a comprehensive analysis of clinical information, gut microbiota data, and antifungal susceptibility phenotypes of intestinal/blood C. parapsilosis isolates from over 200 transplant patients. They found that among the 29 patients who developed gastrointestinal colonization with C. parapsilosis during transplantation, those harboring heterogeneous drug-resistant strains had a significantly higher risk of breakthrough fungemia compared to patients with only susceptible strains colonizing their guts.
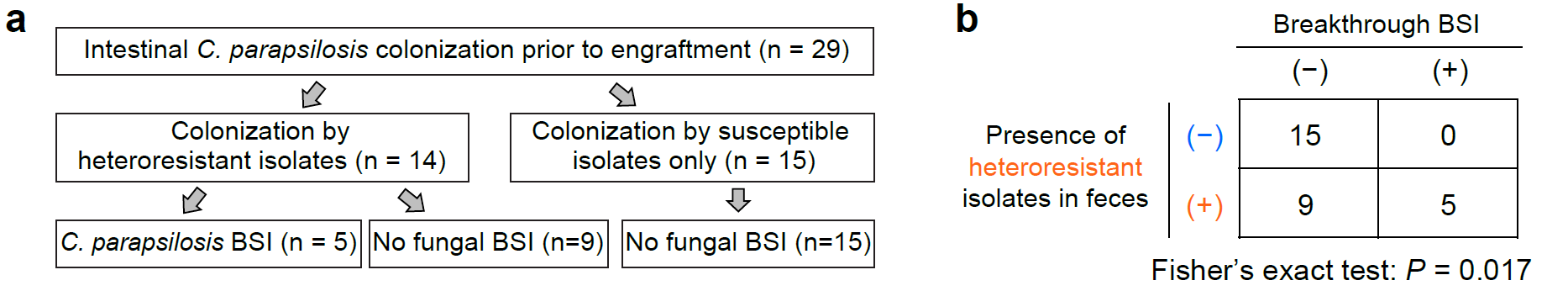
Figure 3. Heterogeneous drug-resistant strains significantly increase the risk of breakthrough fungemia in transplant patients
To explore the prevalence of heterogeneous resistance to micafungin among clinical strains, the authors tested the antifungal susceptibility phenotypes of 219 C. parapsilosis isolates from different medical institutions in the United States, China, France, and Germany against micafungin and performed deep whole-genome sequencing on all isolates. The results revealed that heterogeneously resistant strains to micafungin were found in clinical isolates from various countries, suggesting that this clinically significant drug resistance phenotype may affect the efficacy of echinocandins for the prevention/treatment of fungal infections across a broader geographical range.
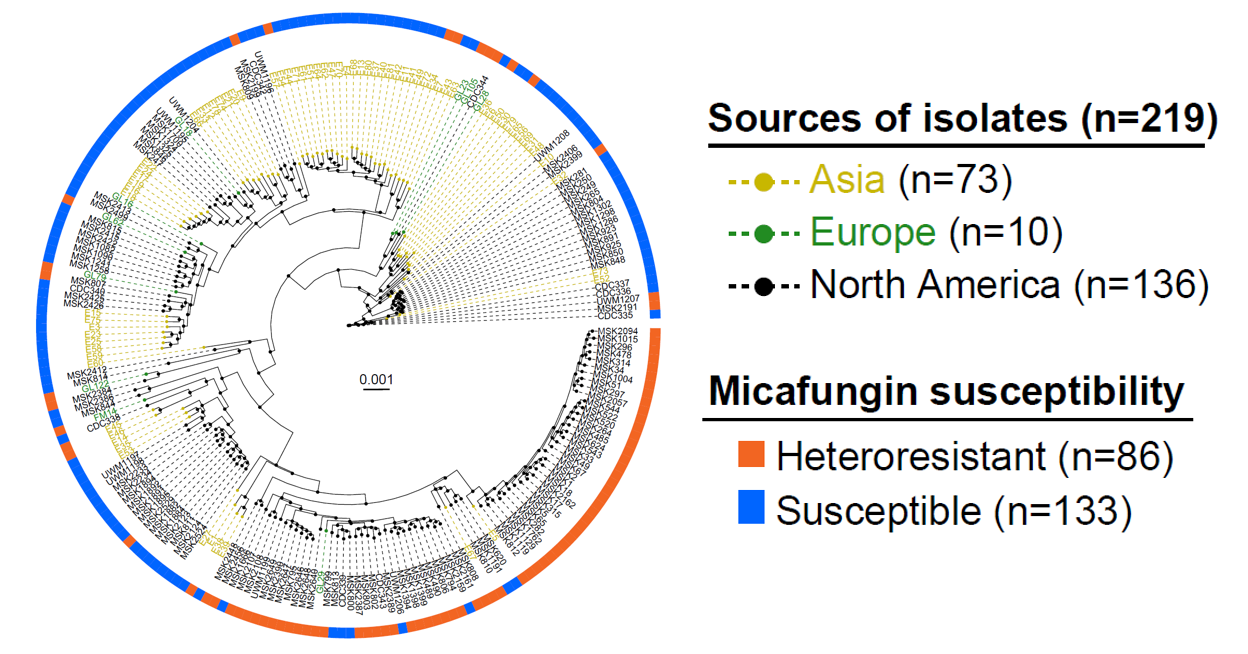
Figure 4. Heterogeneous drug-resistant phenotypes are prevalent in clinical strains from different countries
To precisely characterize the genomic features of Candida parapsilosis and analyze their correlation with heterogeneous drug resistance phenotypes, Dr. Liao Chen, a co-first author of this study, developed a dual-parameter copy number variation (CNV) characterization method based on open reading frame (ORF) integrity and copy number. By integrating the antifungal susceptibility phenotypes and SNV/CNV information of 219 clinical isolates, the authors constructed a machine learning model algorithm that predicts heterogeneous drug resistance phenotypes based on no more than 10 genomic features and performed concept validation. This provides a foundation for the future development of rapid detection methods for heterogeneous drug resistance applicable in clinical settings.
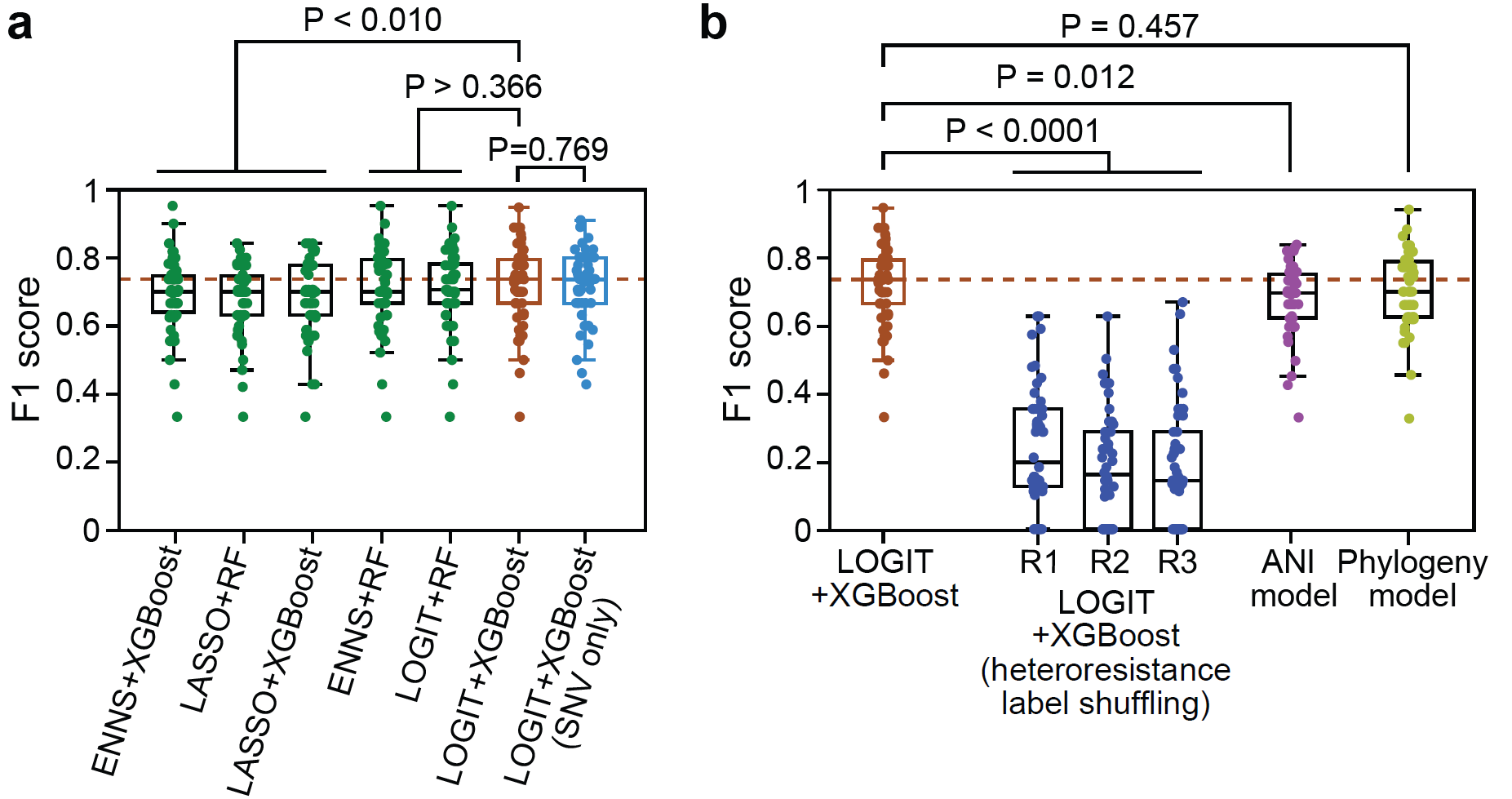
Figure 5. Development of a heterogeneous drug resistance detection method through machine learning strategies
Dr. Zhai Bing from the Institute for Synthetic Biology of the SIAT, and Professor Tobias Hohl from Memorial Sloan Kettering Cancer Center, along with Professor David Weiss from Emory University, are the co-corresponding authors of this paper. Dr. Zhai Bing, Dr. Liao Chen from Memorial Sloan Kettering Cancer Center, Dr. Siddharth Jaggavarapu from Emory University, and Dr. Tang Yuanyuan from the SIAT, are the co-first authors. This study was supported by funding from the National Key R&D Program of China, Ministry of Science and Technology.
Authors:
Dr. Zhai Bing is a researcher and doctoral supervisor at the Institute for Synthetic Biology of the SIAT. Zhai is a national key talent project expert (youth) and the chief scientist of a national key R&D program youth project, whose recent research achievements have been published as first or corresponding author (including co-authorship) in journals such as Nature Medicine (2020 & 2024), Nature Microbiology (2021), and Current Opinions in Microbiology (2021 & 2023). The research team is dedicated to precisely analyzing the adaptive evolution patterns of pathogenic fungi like Candida albicans and Candida parapsilosis during human microbiota colonization and their interactions with commensal bacteria at the same ecological niche, employing synthetic biology techniques to modify microbial community functions, thereby developing new methods for preventing and treating fungal infections.
The research team is always open to postdoctoral researchers, master’s, and doctoral students interested in pathogenic fungal infections, human microbiome interactions, microbial genetics/genomics, etc.
Laboratory homepage: https://zhailabs.wordpress.com/
Dr. Liao Chen currently works at Memorial Sloan Kettering Cancer Center and will join Dartmouth College (Ivy League) as a Tenure-Track Assistant Professor in the Department of Microbiology and Immunology in July 2025, whose research team will combine computational biology, microbiology, and metabolomics to study the metabolic activities and ecological patterns of the human microbiome (bacteria, fungi). Dr. Liao Chen's research has been published as first author (including co-authorship) in top academic journals such as Nature Microbiology, Nature Medicine, Nature Communications, PNAS, eLife, and The ISME Journal, and has received funding from the NIH K99/R00 program.
The research team is always open to postdoctoral researchers, master’s and doctoral students, laboratory assistants, and welcomes visiting scholars/students for exchanges.
Laboratory homepage: https://liaolabatdartmouth.github.io