
On May 8th, the research team led by Ma Yingxin from the Institute of Synthetic Biology of the SIAT, in collaboration with the team led by Ma Lixin from Hubei University and Doctor Yin Wen, published a supplementary cover paper titled "Ultra-Sensitive Detection of the SARS-CoV-2 Nucleocapsid Protein via a Clustered Regularly Interspaced Short Palindromic Repeat/Cas12a-Mediated Immunoassay" in the journal ACS Sensors. This study coupled ELISA, RCA, and the CRISPR/Cas12a systems to develop an ultra-sensitive detection method for the SARS-CoV-2 nucleocapsid protein. The limit of detection (LOD) was as low as 1 fg/mL, four orders of magnitude higher in sensitivity compared to commercial ELISA kits (LOD: 5.7 × 104 fg/mL). This method exhibits high specificity and sensitivity, which can accurately detect SARS-CoV-2 pseudoviruses and clinical samples, providing a new strategy for the ultra-sensitive immunoassay of protein biomarkers.

Screenshot of the article online
Link: https://doi.org/10.1021/acssensors.4c00432
Immunoassays are a common class of bioanalytical techniques that detect and quantify target molecules based on antigen-antibody reactions in biological samples, which are then converted into output signals through reactions with other substances. Among immunoassay methods, enzyme-linked immunosorbent assay (ELISA) is the most commonly used technique for multi-component analysis due to its high throughput and simple operation. However, conventional ELISA has limitations in detecting low-abundance biomarkers. In recent years, clustered regularly interspaced short palindromic repeats (CRISPR) have been widely applied in molecular diagnostics. After CRISPR RNA (crRNA) binds to target DNA, it can activate the cis-cleavage activity of Cas12a, specifically cleaving the target double-stranded DNA and non-specifically cutting free single-stranded DNA in the environment, producing a fluorescent signal. The research team combined the highly sensitive CRISPR/Cas12a system with rolling circle amplification (RCA) and coupled it with ELISA to effectively address the issue of detecting low-abundance biomarkers using ELISA.
This study successfully developed an ultrasensitive immunoassay for detecting the SARS-CoV-2 nucleocapsid (N) protein. As shown in Figure 1, after the SARS-CoV-2 N protein is captured by antibodies coated on a well plate, it sequentially binds to primary antibodies, secondary antibodies, streptavidin, and biotin-conjugated primers, forming a sandwich-like immune complex. The padlock probe is cyclized by a ligase and then RCA is initiated under the action of polymerase to produce single-stranded DNA. After CRISPR/Cas12a binds to the single-stranded DNA, it activates its trans-cleavage activity, cleaving the FAM-ssDNA-BHQ1 probe and releasing a fluorescent signal, thereby achieving ultrasensitive detection of the SARS-CoV-2 N protein.
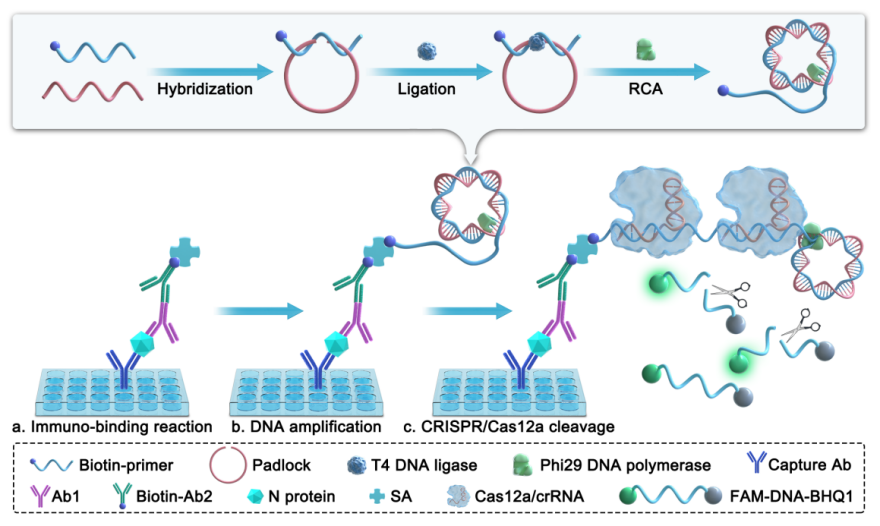
Figure 1 Schematic diagram of the CRISPR/Cas12a-mediated immunoassay for detecting SARS-CoV-2 N protein. (a) The capture antibody captures the N protein, which is then bound by the primary antibody, biotinylated secondary antibody, and streptavidin to form an immune complex; (b) Under the action of phi29 DNA polymerase, the biotinylated primer initiates the RCA reaction using the circularized padlock as a template, producing long ssDNA; (c) CRISPR/Cas12a recognizes the long ssDNA strand and trans-cleaves the free FAM-ssDNA-BHQ1.
In this study, the team first evaluated the feasibility of RCA and the CRISPR/Cas12a system. The experimental results showed that the CRISPR/Cas12a system could recognize RCA products and produce fluorescent signals. Subsequently, the team developed an immunoassay combining ELISA, RCA, and CRISPR/Cas12a. The detection results showed that as the concentration of the N protein gradually increased, the fluorescent signal gradually rose, and there was a good linear relationship between the logarithm of the concentration and the fluorescent signal (Figures 2A-C), with a limit of detection (LOD) as low as 1 fg/mL, significantly outperforming commercial ELISA kits in terms of detection sensitivity by 4-5 orders of magnitude (Figure 2D). Moreover, this method demonstrated excellent detection specificity for the SARS-CoV-2 N protein (Figure 3A). When comparing this method with commercial kits for the detection of SARS-CoV-2 pseudovirus, the results indicated that our method could accurately differentiate between positive and negative samples, while the commercial kits could not achieve this (Figures 3B-C). Additionally, when analyzing clinical samples of COVID-19 using this method, the results achieved 100% accuracy (Figure 3D).
In summary, the team successfully constructed an ultrasensitive immunoassay method coupling ELISA, RCA, and the CRISPR/Cas12a systems, achieving ultrasensitive detection of the SARS-CoV-2 N protein. CRISPR/Cas12a-mediated immunoassays can significantly enhance detection sensitivity, providing an important technical means for ultra-early and precise diagnosis of infectious diseases and addressing key needs in ultrasensitive detection in both biological research and clinical diagnostics.
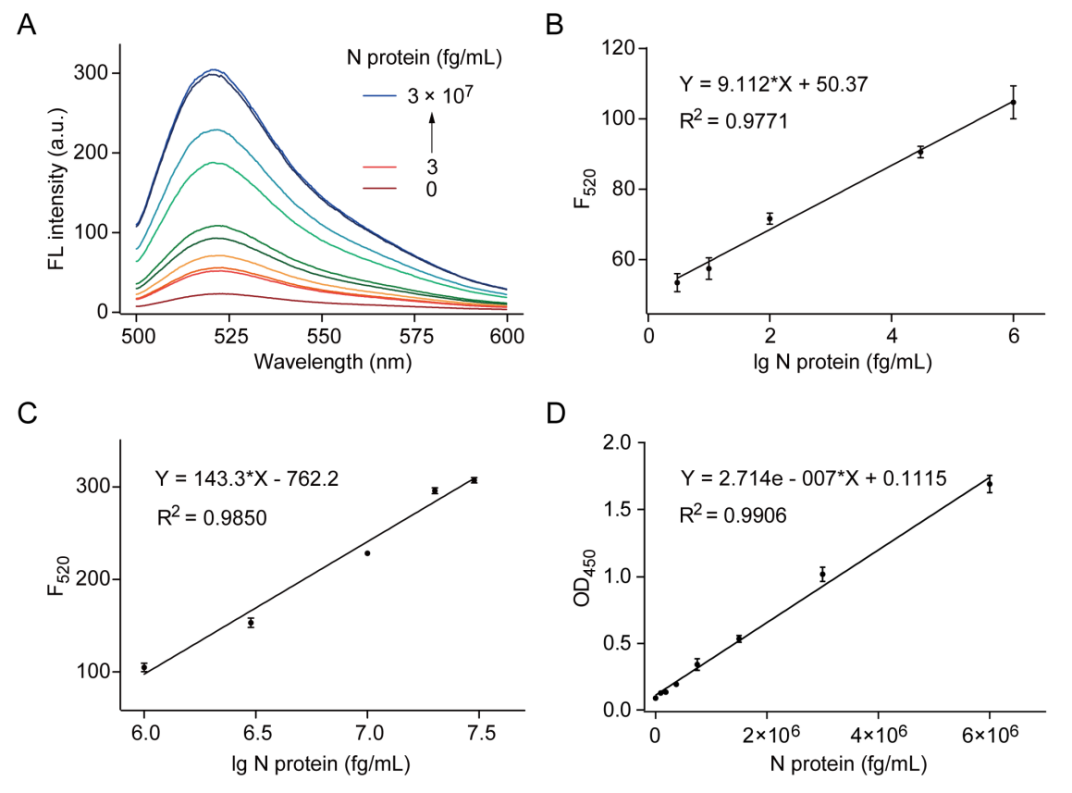
Figure 2 CRISPR/Cas12a-mediated immunoassay for N protein detection. (A) Fluorescence spectrum of the reporter probe response at different concentrations of N protein in the CRISPR/Cas12a-mediated ELISA experiment; (B) and (C) show the linear relationship between the logarithm of N protein concentration and fluorescence intensity at 520 nm, with concentration ranges of 3 fg/mL to 1 × 106 fg/mL and 1 × 106 fg/mL to 3 × 107 fg/mL, respectively; (D) Linear relationship of a commercial ELISA kit for N protein detection (1 × 105 fg/mL to 6 × 106 fg/mL).
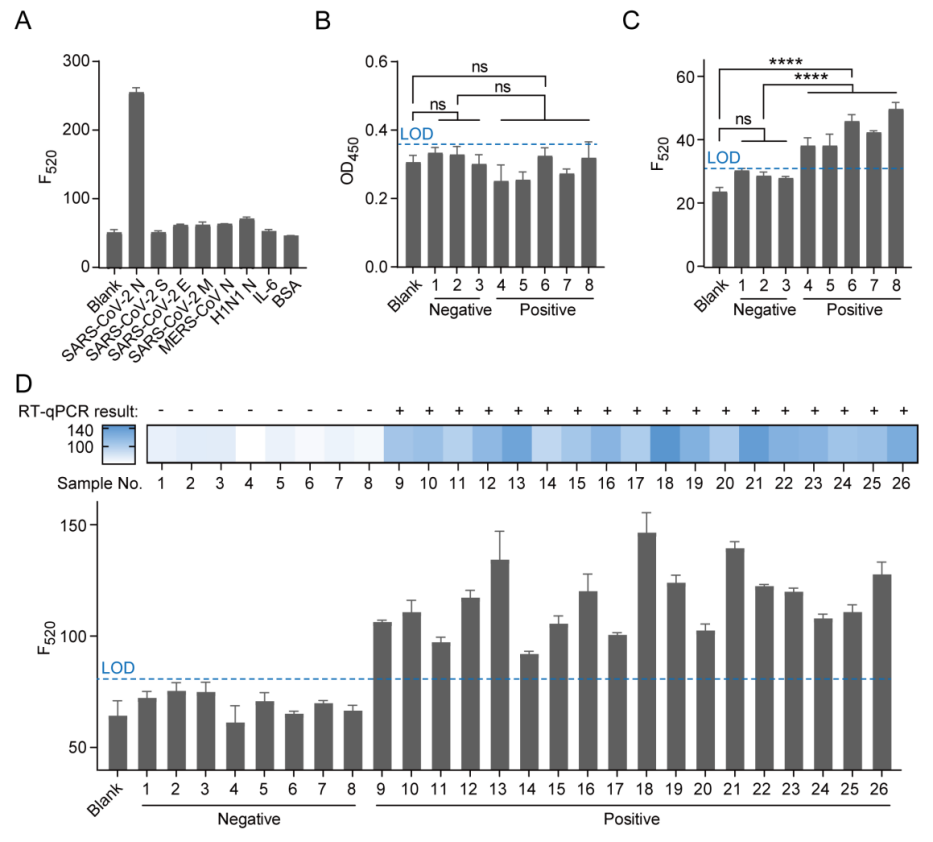
Figure 3 Selectivity study and virus sample detection. (A) Selectivity study of the SARS-CoV-2 N protein. The concentration of N protein is 5 ng/mL, while the concentrations of other proteins are 50 ng/mL each; (B) Commercial ELISA kit and (C) CRISPR/Cas12a-mediated immunoassay for detecting SARS-CoV-2 pseudovirus samples. Samples 1 to 3 are negative controls, while samples 4 to 8 are positive controls; (D) Analysis of clinical samples from COVID-19 patients. Samples 1-8 are negative, while samples 9-26 are positive. LOD = blank + 3 × standard deviation.
Doctor Yin Wen from Hubei University, Li Leyao, a co-trained master's student with the SIAT, and Yang Yang, a researcher with Shenzhen Third People’s Hospital, are co-first authors of the paper. Researchers Ma Yingxin and Mao Guobin from the Institute of Synthetic Biology of the SIAT, are the corresponding authors of the paper. Researchers Liang Ruijing from the SIAT, Ma Lixin from Hubei University, and Dai Junbiao from the Shenzhen Institute of Agricultural Genomics, Chinese Academy of Agricultural Sciences, participated in this research.
This project was supported by different programs such as the National Natural Science Foundation of China, the Ministry of Science and Technology, the Chinese Academy of Sciences, Guangdong Province, Shenzhen City, and the Shenzhen Institute of Synthetic Biology.