
On May 8, 2024, the research team led by Researcher Gan Haiyun from the Institute of Synthetic Biology of the SIAT, and the Shenzhen Institute of Synthetic Biology, along with Professor Yu Chuanhe from the Hormel Institute at the University of Minnesota and Professor Han Junhong's team from the State Key Laboratory of Biotherapy and Frontiers Science Center for Disease-related Molecular Network at West China Hospital, Sichuan University, published an article titled "DNA polymerase delta governs parental histone transfer to DNA replication lagging strand" in PNAS (Proceedings of the National Academy of Sciences). The study discovered that DNA polymerase delta (Pol δ) acts as a key companion protein during DNA replication, transferring parental histones to the lagging strand. This ensures that epigenetic information carried by post-translational modifications of parental histones is faithfully transmitted to progeny cells. This finding reveals the intricate process by which Pol δ promotes the inheritance of parental histones during DNA replication on the lagging strand. Pol δ is known to be involved in DNA damage repair, but its function was previously unknown, suggesting that histone recycling might play a role in DNA damage repair.
Screenshot of the article online
Link: https://www.pnas.org/doi/10.1073/pnas.2400610121
On March 8th, the research team led by Researcher Gan Haiyun and Professor Yu Chuanhe from the Hormel Institute at the University of Minnesota published an article titled "Defective transfer of parental histone decreases frequency of homologous recombination by increasing free histone pools in budding yeast" in Nucleic Acids Research (NAR). The study clarified the genetic and epigenetic consequences arising from the disruption of parental histone H3-H4 tetramer transfer. This discovery revealed that the correct transfer of parental histones during DNA replication is not only crucial for maintaining the stability of chromatin structure, but also that any decrease in homologous recombination activity due to defects in parental histone transfer can adversely affect cell health.

Screenshot of the article online
Link: https://doi.org/10.1093/nar/gkae205
DNA replication and histone transfer are two closely linked processes in living organisms, together ensuring the accurate transmission of genetic information and the normal functioning of cells. During DNA replication, the parental nucleosome structure ahead of the replication fork needs to be disassembled, while histones from the parental nucleosomes are transferred to the newly synthesized daughter strands behind the replication fork, including both the leading and lagging strands, where they reassemble into nucleosomes, facilitating the transmission and inheritance of epigenetic information. In recent years, Researcher Gan Haiyun and collaborators developed a technology capable of specifically detecting proteins bound to nascent DNA during DNA replication (enrichment and sequencing of protein-associated nascent DNA, eSPAN) and discovered that DNA polymerase Pol ε, while synthesizing DNA, has subunits Pole3 and Pole4 responsible for transferring parental histones to the leading strand (Science, 2018). Mcm2 (a subunit of the Mcm2-7 helicase) transfers parental histones to the lagging strand of replicating DNA through Polα (Molecular Cell, 2018). However, MCM2 itself is located on the leading strand, and Polα can only synthesize RNA primers and approximately 30 nt of DNA sequence, indicating that the existing pathways alone cannot complete histone recycling on the lagging strand, which has remained a puzzle for researchers.
After extensive screening and experimental validation, the research team led by Gan Haiyun and collaborators discovered that the absence of the non-essential subunit Pol32 of DNA polymerase δ leads to the predominant transfer of parental histone H3-H4 to the leading strand during replication. Further biochemical analysis demonstrated that Pol32 can bind to histone H3-H4 both in vivo and in vitro (Figure 2).
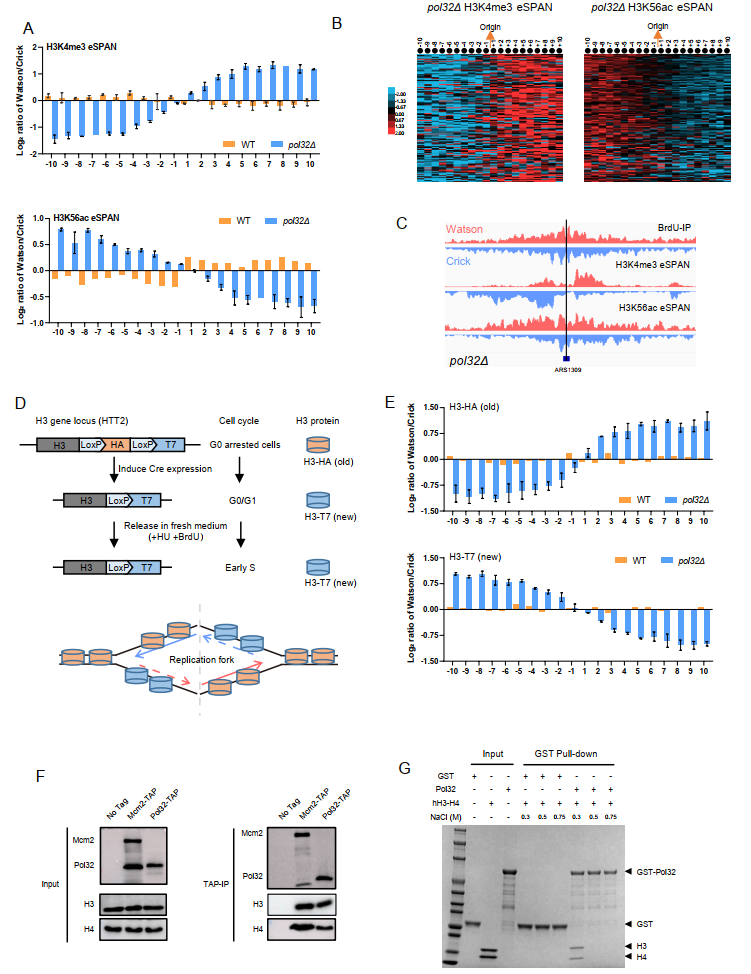
Figure 2. Pol32-mediated transfer of histone H3-H4 to the lagging strand
Researchers also discovered that by mutating the histone H3-H4 binding domain of Mcm2, the interaction between Pol32 and parental histone H3-H4 was disrupted, further confirming the close connection between Mcm2 and Pol32. Integrating these findings, the DNA polymerase δ subunit Pol32 has been identified as a key histone chaperone downstream of Mcm2, capable of mediating the transfer of parental histones to the lagging strand during DNA replication (Figure 3). This discovery not only reveals a critical link in the mechanism of parental histone transfer, but also provides new directions for future research on the transmission of genetic and epigenetic information.
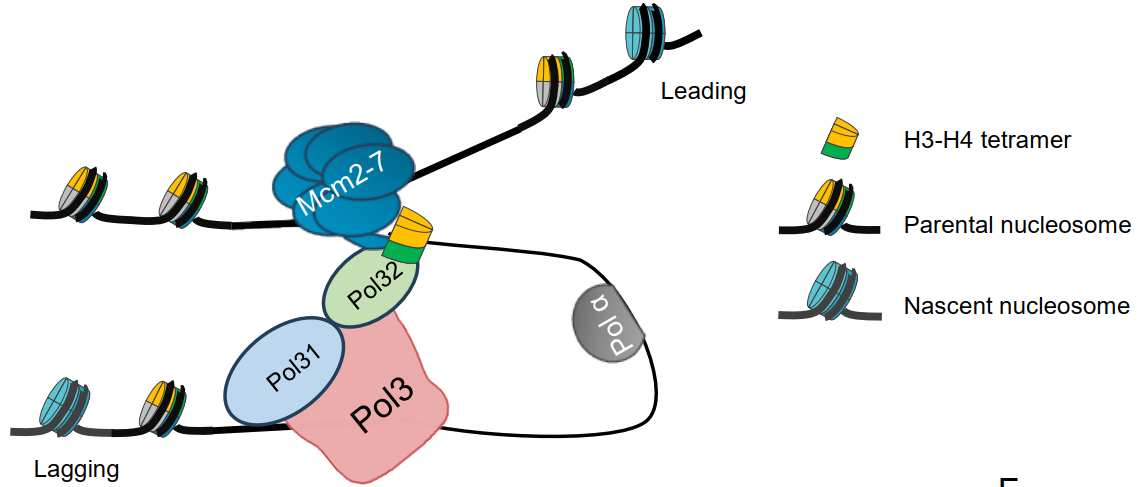
Figure 3. Interaction of Mcm2, H3-H4, and Pol32 at the replication fork
Assistant researcher Tian Congcong, assistant researcher Zhou Jiaqi, research assistant Zhang Ziwei from the SIAT, and Ph.D. student Zhang Qin from Sichuan University, together with postdoctoral fellow Jia Jing from the University of Minnesota, are co-first authors of the paper. This research was supported by multiple programs including the National Key R&D Program for Synthetic Biology, the National Natural Science Foundation of China, Strategic Priority Research Program of the Chinese Academy of Sciences, General Programs and Youth Programs of the National Natural Science Foundation of China, Guangdong Provincial Natural Science Fund for Distinguished Young Scholars, Shenzhen Institute of Synthetic Biology Research Plan, and Shenzhen Medical Research Foundation.
Expert Review
Professor Zhu Weiguo from Shenzhen University
In recent years, studies have shown that the inheritance of parental histones is crucial for epigenetic remodeling in daughter cells, maintenance of differentiation characteristics, and even individual development. The coupling of DNA replication with the transfer of parental histones ensures the stable inheritance of the epigenome. Previously, Gan Haiyun and collaborators found that the helicase subunit MCM2 and DNA polymerase polα were responsible for the transfer of parental histones to the lagging strand. However, since MCM2 is located on the leading strand and polα, as a primase, can only synthesize the RNA primer and about 30 nt of DNA sequence required for lagging strand replication, it is clear that they cannot complete the entire process of parental histone recycling on the lagging strand. This has been a puzzle for some time. In this study, led by Gan Haiyun's team, it was discovered that the non-essential subunit pol32 of DNA polymerase polδ can act as a histone chaperone to transfer parental histones to the lagging strand in yeast cells. This research completes the last piece of the puzzle in the pathway of histone recycling coupled with DNA replication, perfects the theory of DNA replication-coupled histone recycling, and finally unifies DNA replication with chromatin duplication. Furthermore, there is evidence suggesting that polδ may be involved in the DNA damage repair process. Gan Haiyun and collaborators' recent NAR article also found that defects in parental histone transfer affect the frequency of homologous recombination. These studies not only fill the previous knowledge gap regarding the mechanism of histone recycling, but also provide a new perspective on chromatin repair during the DNA damage repair.
PI and Group Profile
Dr. Gan Haiyun, researcher with the Institute of Synthetic Biology of the SIAT, and doctoral supervisor. In the past five years, Gan’s research achievements have been published in renowned journals such as Science, Nature Medicine, Nature Genetics, Molecular Cell, Nature Communications, Genes & Development, PNAS, eLife, The EMBO Journal, and Nucleic Acids Research, accumulating over 3400 citations.
The research group mainly focuses on studying the mechanisms of epigenome information transmission and its regulatory roles in tumorigenesis and drug resistance, pluripotent cells (2C), cell aging, extrachromosomal DNA (ecDNA), synthetic epigenetic reprogramming, DNA replication initiation molecular complexes, artificial cells, etc., using a combination of "wet" and "dry" experiments in systems biology and synthetic biology.
The group is currently recruiting 1-2 postdoctoral researchers and 2-3 research assistants with backgrounds in any of the following fields: epigenomics, synthetic biology, bioinformatics, tumor biology, cell biology. Interested candidates are welcome to send their CVs to hy.gan@siat.ac.cn, with the subject indicating "Application Position - School Name - Major - Full Name."