
Esophageal cancer ranks among the top in both incidence and mortality rates globally, with over 50% of cases occurring in China, where approximately 90% are pathologically classified as esophageal squamous cell carcinoma (ESCC). ESCC exhibits extensive tumor heterogeneity in terms of genomic alterations and the tumor microenvironment. The causes of this heterogeneity in ESCC remain unclear. A comprehensive analysis of the characteristics and mechanisms underlying tumor heterogeneity can guide drug development and clinical treatment for ESCC. On April 23, 2024, a collaborative team led by Academician Zhan Qimin and Researcher Cui Yongping from the Medical Center for Shenzhen Peking University/Hong Kong University of Science and Technology/Shenzhen Bay Laboratory’s Tumor Research Institute, along with Researcher Hu Zheng from the Shenzhen Institute of Advanced Technology, Chinese Academy of Sciences, Professor Zhihua Liu from the Cancer Hospital of the Chinese Academy of Medical Sciences, and the Shanxi Provincial Cancer Hospital team, published their latest research findings online in the journal National Science Review under the title "Immune-tumor interaction dictates spatially directed evolution of esophageal squamous cell carcinoma." This study used multi-omics technologies to systematically characterize the spatial heterogeneity map of ESCC. Based on the interactions among environmental factors (alcohol consumption), the tumor microenvironment (immune response), and spatial clonal evolution (tumor), the study proposed a new model for spatially directed tumor evolution. It also identified a novel gene associated with ESCC, PREX2, providing new insights into the pathogenesis of ESCC.

Screenshot of the article published online
Link to the article: https://academic.oup.com/nsr/advance-article/doi/10.1093/nsr/nwae150/7656974?searchresult=1
This study conducted multi-omics sequencing on 507 spatially multiplexed samples from 103 ESCC patients in China's Taihang Mountain area, including whole exome sequencing, transcriptome sequencing, and 850K methylation array sequencing. Each sample had clear spatial location information, originating from the "upper-lower-left-right-middle", five points of the tumor. Based on the large-scale omics data, we performed an in-depth analysis of the interactions among mutation-driven genes, clonal evolution patterns, immune microenvironment, and living environment. The spatial heterogeneity map of ESCC was elucidated, and a targeted evolution model was proposed for esophageal squamous cell carcinoma and potential therapeutic targets. This provides important theoretical support for understanding the tumor evolution mechanism of ESCC in the Chinese population and clinical sampling.
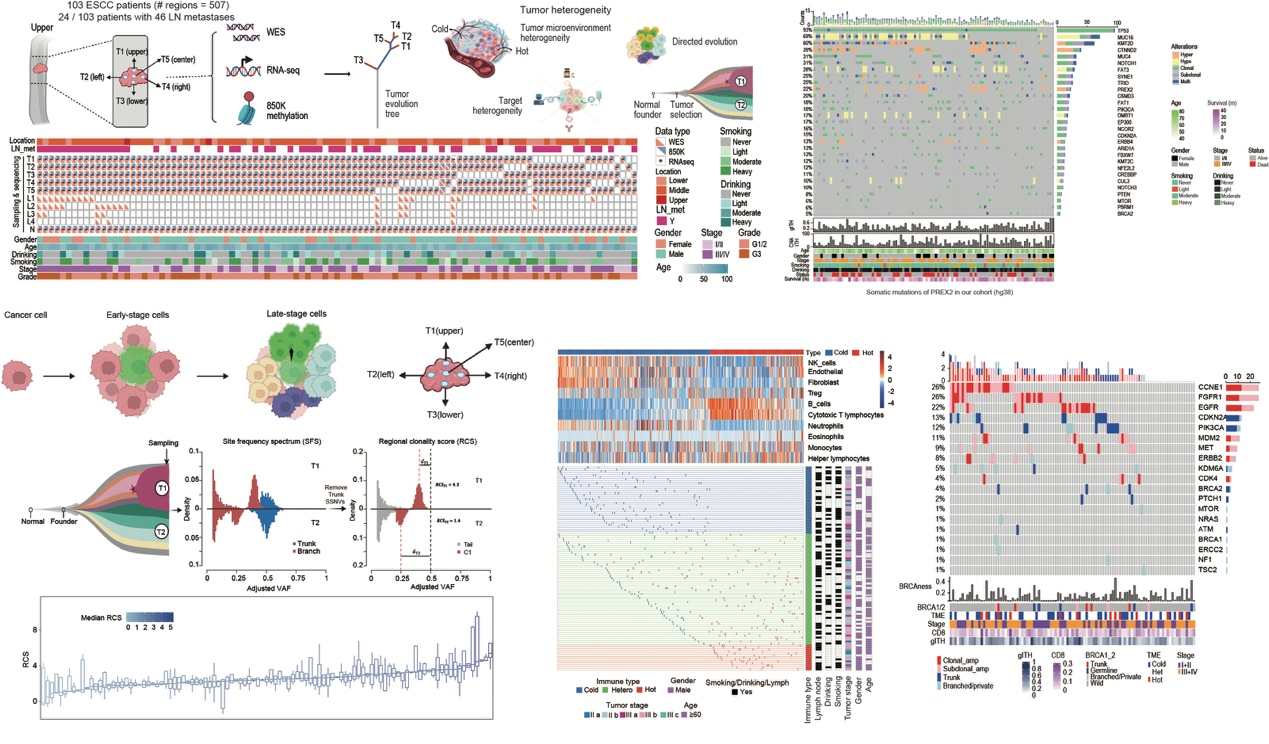
Figure. Spatial Heterogeneity and Directed Evolution Patterns of Esophageal Squamous Cell Carcinoma (ESCC)
Firstly, the researchers constructed a spatial sampling and multi-omics dataset for ESCC to explore and identify key driver genes at the patient level. They found that the mutation frequency of most oncogenes was higher at the patient level compared to the sample level. Chromosomal arm changes and TP53 mutations primarily occurred in the early stages, while most point mutations, such as those in NFE2L2, appeared in the later stages. Notably, a considerable portion of copy number alteration driver genes occurred early, suggesting that copy number alterations might be a key factor driving early-stage esophageal squamous cell carcinoma. Additionally, this part of the study emphasized the methylation changes in ESCC, including genes driven by hypermethylation like KMT2D and PREX2, with PREX2 being a newly identified hypermethylation driver gene. Functional experiments were also conducted on the PREX2 gene, revealing its feasibility as a potential target.
Then, by constructing tumor evolution trees and performing dynamic evolution analysis, the researchers discovered that tissues from the upper part of the tumor had higher similarity to the tumor center in terms of both the genome and transcriptome levels compared to other locations. This suggests that subclones originating from the tumor center are more likely to expand towards the upper esophagus. Based on these results, the paper proposed a model of spatially directed evolution for tumors. To validate this evolution model, the researchers analyzed the immune microenvironment of ESCC and the immune editing capabilities of different subclones. They found that these data still supported the evolution model. The study also found that the immune editing score (immune editing score) was significantly associated with the prognosis of ESCC, indicating it could serve as a prognostic marker for ESCC. In contrast, tumor TMB or CD8+ cells were not significantly correlated with the overall prognosis of ESCC, a finding that was validated in multiple independent datasets. This indicates that the immune microenvironment features of ESCC differ from those of other human tumors. To further elucidate the reasons behind this evolution model, the authors analyzed the relationship between patients' lifestyle habits and clonal evolution. They discovered that ESCC patients who consumed alcohol were more likely to exhibit a spatially directed evolution pattern, revealing the mechanism of alcohol-driven tumor clonal evolution and microenvironment interaction.
Finally, to study the heterogeneity of therapeutic targets, the article collected mutations and amplification-driven potential targets involved in the oncoKB database. A total of 19 target genes involving 152 actionable mutation sites were identified, with 49.3% of mutations being heterogeneous. Common amplified genes included EGFR and ERBB2. From the perspective of previous clinical treatment and research, some patients with BRCAness tumors may benefit from platinum-based chemotherapy. Since platinum-based chemotherapy is often used before or after surgery for ESCC, these results suggest that ESCC patients with high BRCAness signals may benefit from this treatment. Specifically, among 103 patients, 14 (13.5%) carried BRCA1/2 germline or somatic mutations, and BRCAness patients showed better survival outcomes. This conclusion was also validated in multiple datasets. These results indicate that ESCC patients with high BRCAness may be suitable for specific treatment strategies.
In summary, this study integrated multi-omics data from multiple tumor sites to comprehensively demonstrate the multi-level heterogeneity of ESCC and its formation. The proposed spatially directed evolution model helps unravel the pathogenesis and clonal evolution mechanisms of ESCC in the Chinese population, providing important theoretical support for the precise prevention and treatment of ESCC.
The co-corresponding authors of the paper are Academician Zhan Qimin and Cui Yongping from the Shenzhen Peking University/Hong Kong University of Science and Technology Medical Center/Shenzhen Bay Laboratory’s Tumor Research Institute, Researcher Hu Zheng from the SIAT, and Professor Liu Zhihua from the Cancer Hospital of the Chinese Academy of Medical Sciences. Dr. Zhou Yong from the Shenzhen Peking University/Hong Kong University of Science and Technology Medical Center/Shenzhen Bay Laboratory’s Tumor Research Institute, Dr. Mo Shanlan from the SIAT, Assistant Professor Cui Heyang from the Department of Surgery, Chinese University of Hong Kong, Doctors Sun Ruifang, Zhuang Xiaofei from Shanxi Provincial Cancer Hospital, and Associate Researcher Zhang Weimin from the Shenzhen Peking University/Hong Kong University of Science and Technology Medical Center/Shenzhen Bay Laboratory’s Tumor Research Institute are the co-first authors of the paper. This project was supported by the Guangdong Fund for Major Basic and Applied Research Projects, Shenzhen Medical Research Special Fund, National Key R&D Program, Independent Major Project of Shenzhen Bay Laboratory, Shanxi Province Higher Education Fund, "1331 Project" Fund and Science and Technology Innovation Team Special Fund, National Natural Science Foundation, Shenzhen Institute of Synthetic Biology, etc.