
On March 15, 2024, the Zhao Qiao team from the Center for Synthetic Genomics at the Shenzhen Institute of Advanced Technology, Chinese Academy of Sciences, published a research paper titled "6-Phosphogluconate dehydrogenase 2 bridges the OPP and shikimate pathway to enhance aromatic amino acid production in plants" in SCIENCE CHINA Life Sciences. This work reveals that PGD2 can couple the oxidative pentose phosphate (OPP) pathway and the shikimate pathway to regulate the synthesis of aromatic amino acids and their derivatives from a metabolic flux perspective.

Original Link: https://www.sciengine.com/SCLS/doi/10.1007/s11427-024-2567-4
Aromatic amino acids (AAAs) are primarily synthesized through the shikimate pathway in plant plastids. They are not only the basic building blocks of proteins but also precursors for many important secondary metabolites, such as auxins, lignins, and flavonoids. In addition to synthesizing a baseline level of aromatic amino acids for protein biosynthesis, plants must also regulate the carbon flux toward aromatic amino acid synthesis in response to changes in environmental conditions to maintain the production of downstream aromatic metabolites. Therefore, controlling the appropriate level of aromatic amino acid synthesis is critical for plant growth, reproduction, and stress resistance.
The oxidative pentose phosphate (OPP) pathway exists in all eukaryotes and provides metabolic intermediates for the shikimate pathway, directing carbon flux toward the biosynthesis of AAAs and numerous aromatic natural products. Although the basic aspects of the OPP pathway have been elucidated, the details of how this pathway influences AAA synthesis remain unclear.
The Zhao Qiao team previously conducted a forward genetic screening based on an Arabidopsis mutant with reduced aromatic amino acid synthesis, identifying several plants that could restore the mutant phenotype. Further genomic sequencing analysis identified multiple new genes potentially involved in the synthesis and metabolic regulation of aromatic amino acids and their derivatives in Arabidopsis. In this study, the team conducted an in-depth analysis of one of these genes, which encodes 6-phosphogluconate dehydrogenase PGD2. A single nucleotide mutation in the PGD2 exon changes the 63rd amino acid from glutamic acid (Glu, E) to lysine (Lys, K). The presence of this point mutation partially restores the defect phenotype of adh2.
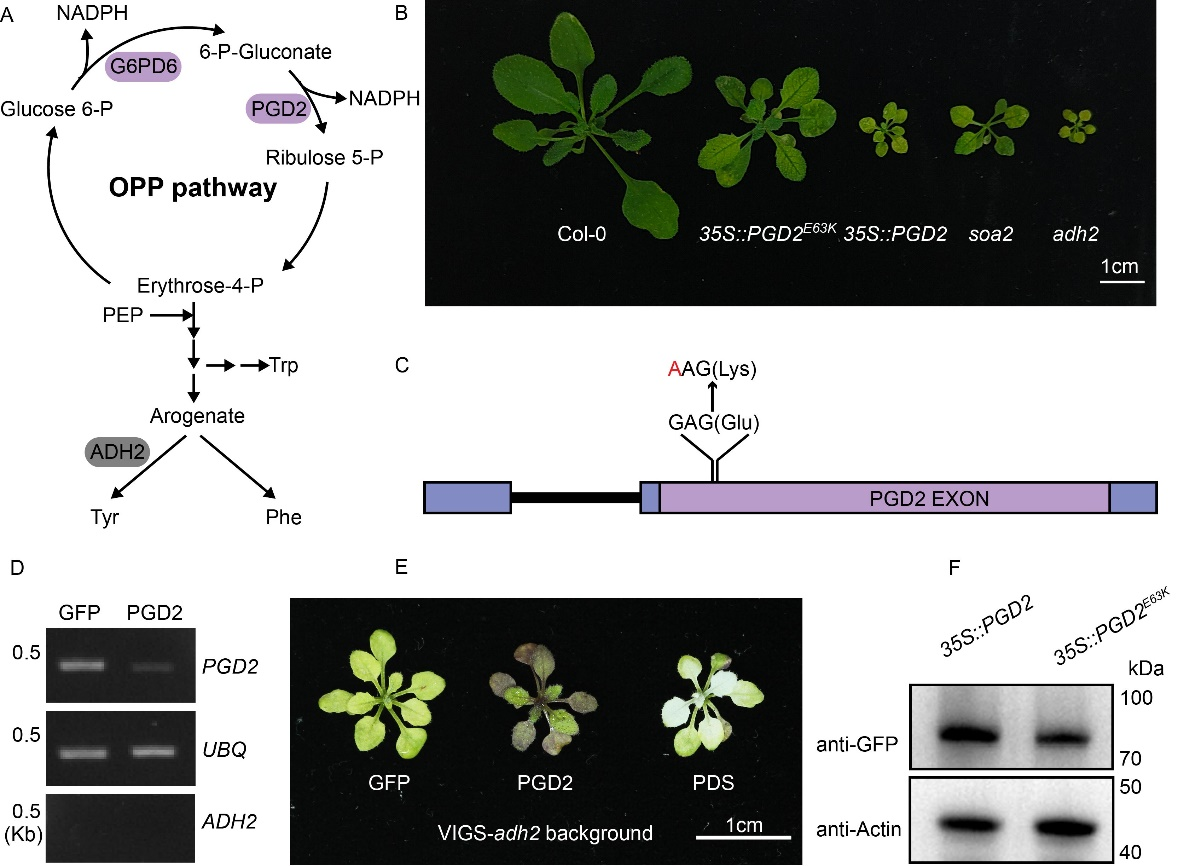
Figure 1 A point mutation in PGD2 restores the growth deficiency phenotype of adh2.
Biochemical analysis showed that the catalytic activity of PGD2E63K is significantly higher than that of PGD2. Based on the natural PGD2 protein sequence, targeted mutations were made to the 63rd amino acid residue of Arabidopsis PGD2, and enzyme activity was further compared. The catalytic activities of PGD2E63R (R, Arg) and PGD2E63K were stronger than those of PGD2E63A (A, Ala) and PGD2, indicating that when the residue at this position is a basic amino acid (K and R), the enzyme activity is stronger than when it is an acidic amino acid (E). Neutral mutations, such as A, had almost no effect on PGD2 enzyme activity.
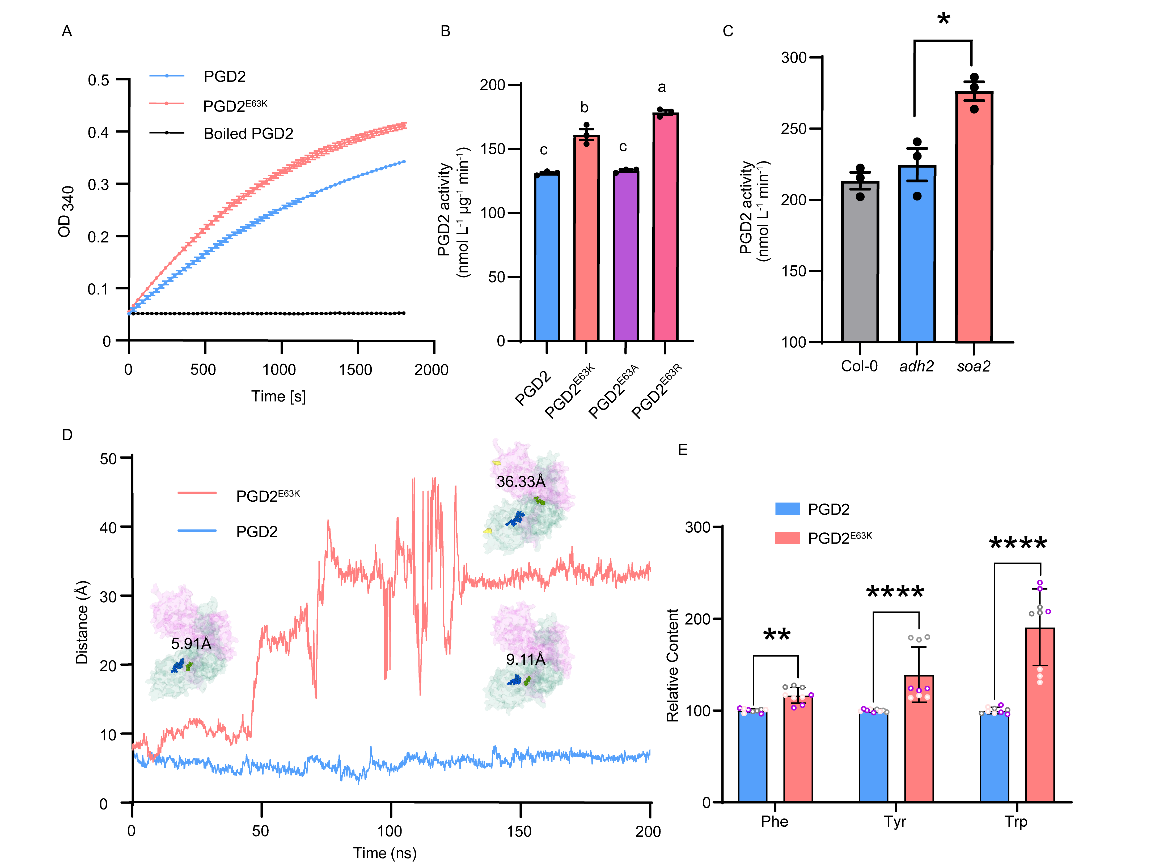
Figure 2 Point mutations enhance the enzymatic activity of PGD2.
Through τ-random accelerated molecular dynamics (τRAMD) calculations, it was found that compared to natural PGD2, the products R5P and NADPH dissociate from PGD2E63K more quickly. This indicates that the point mutation in PGD2 enhances enzyme activity by promoting product release and maintaining sustainable enzyme turnover.
Additionally, quantitative analysis using the Agilent 1290 ultra-high-performance liquid chromatography coupled with a 6470 triple quadrupole mass spectrometer (UPLC-QQQ-MS/MS) showed that the presence of PGD2E63K increased the levels of AAAs and their derivatives—including α-tocopherol, tyramine, 4-hydroxycinnamic acid, pinene, and flavonoids—in plants. Therefore, PGD2 directs carbon flux toward the shikimate pathway, promoting the synthesis of aromatic amino acids and their derivatives.
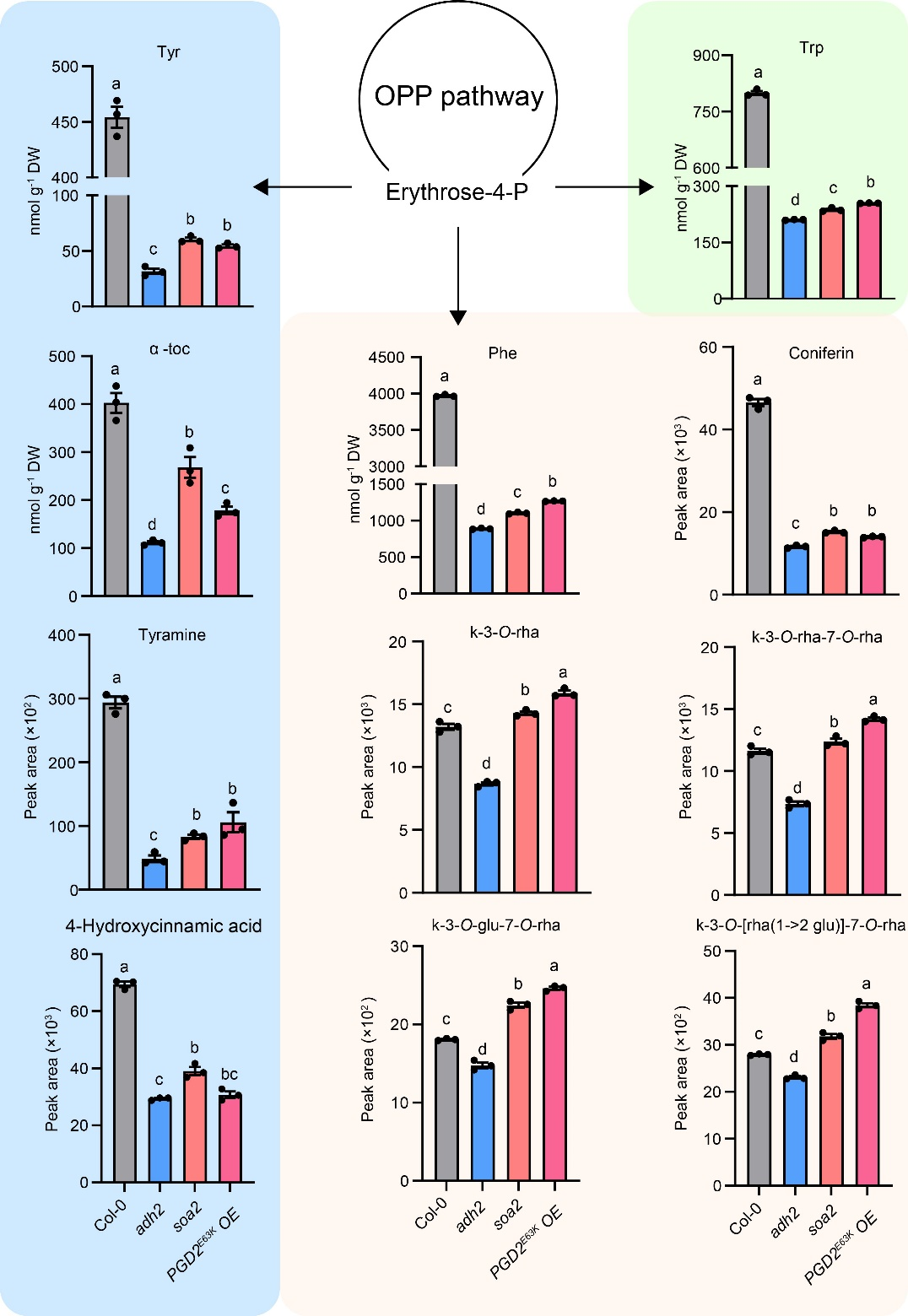
Figure 3 PGD2E63K increases the content of aromatic amino acids and their derivatives in Arabidopsis.
Since the identified point mutation occurs in a relatively conserved amino acid residue, the team performed site-directed mutagenesis and enzyme activity analysis on PGD proteins from bacteria, algae, mosses, gymnosperms, monocots, and dicots. They found that the conclusions drawn from Arabidopsis have a certain universality across different species, where the type of amino acid at this position affects PGD's catalytic activity, with basic amino acids at this position conferring higher reaction efficiency than acidic amino acids.
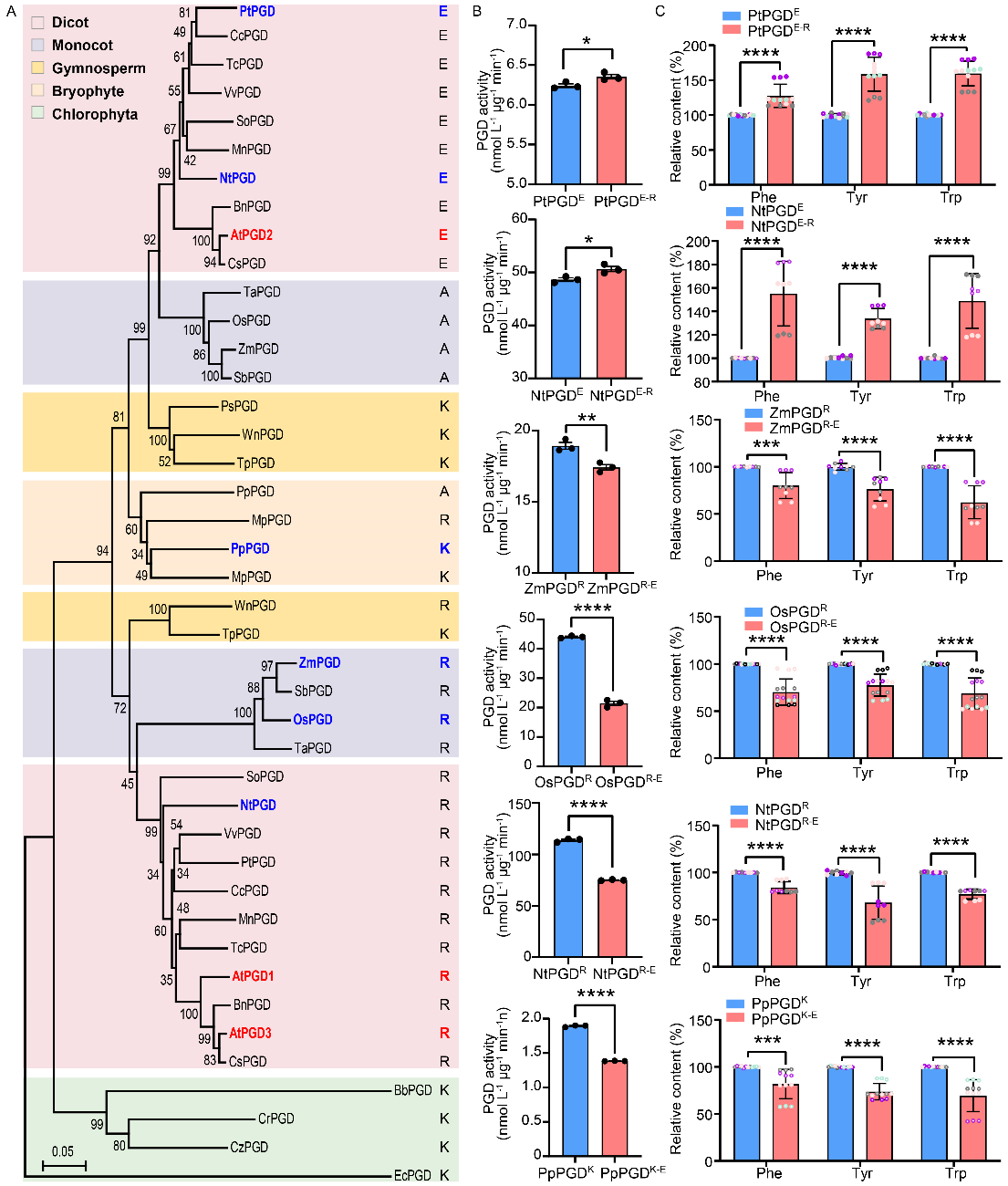
The above work elucidates a new mechanism for the regulation of aromatic amino acid and their derivative synthesis in plants from a metabolic flux perspective, revealing the interaction between PGD2, the OPP pathway, and the shikimate pathway, directing carbon flux toward the biosynthesis of aromatic amino acids. The identified PGD2 mutation site could serve as a potential target for metabolic engineering design to regulate metabolic flux and enhance the synthesis of downstream compounds in the OPP pathway.
Researcher Zhao Qiao and assistant researcher Wu Jie from the Shenzhen Institute of Advanced Technology, Chinese Academy of Sciences, are the corresponding authors of the article. Postdoctoral fellow Tang Qian and graduate student Huang Yuxin are the co-first authors. This research was supported by the National Key Research and Development Program, the National Natural Science Foundation, and the Guangdong Provincial Key Laboratory of Synthetic Genomics, Shenzhen Institute of Synthetic Biology Innovation.