
On March 9, 2024, the research team led by Dai Zhuojun with Shenzhen Institute of Advanced Technology, Chinese Academy of Sciences, published an online research paper titled “Engineering Functional Materials through Bacteria-assisted Living Grafting” in Cell Systems, a sub-journal of Cell. Drawing on the strategy of chemical grafting reaction, this study programmed microorganisms through synthetic biology, achieved the continuous production and in situ grafting of functional proteins, and endowed the materials with diverse biological functions. In the traditional chemical grafting reaction, although the functionalization of macromolecules or their surface can be achieved, the endowed properties are often limited by physical and chemical attributes such as hydrophobicity, pH, temperature sensitivity, in lack of biological functions (such as mismatched DNA binding). In contrast, this study draws on the idea that donor plants can grow and provide multiple scions in plant grafting, and by regulating the continuous growth of engineered bacteria and release of functional protein module, the stability and self-repair ability of grafting response were enhanced, and a series of dynamic adjustable and renewable biological functions were achieved. The research paper was also selected as the cover article of the journal Cell Systems.

Screenshot of the article
Link: https: / / www.sciencedirect.com/science/article/abs/pii/S2405471224000577?via%3Dihub

Cover article of the journal Cell Systems
The team encapsulated engineered bacteria with polymer scaffold containing approximately 20-micron microparticles with active grafting sites; these microorganisms that have been regulated by genetic circuit can grow in the scaffold, sense their own density and release specific functional proteins through autonomous cleavage; the target protein carrying the tag can effectively graft to the active sites in the scaffold to assemble into MELG (Materials Engineered by Living Grafting: MELG) (Figure 1). The system is highly modular: chassis cells providing protein modules, customizable genetic circuits, multiple functional protein modules, and chemical graft reaction, all of which can be adjusted flexibly. Thanks to the continuous supply of grafted proteins by living microorganisms, the system shows high levels of stability and regeneration to external disturbance. On this basis, the team further developed a variety of multi-functional MELG, such as controlled release of proteins in response to biological signals, synthesis of the high-value chemical product olivonic acid, and correction of mismatched DNA sequences.
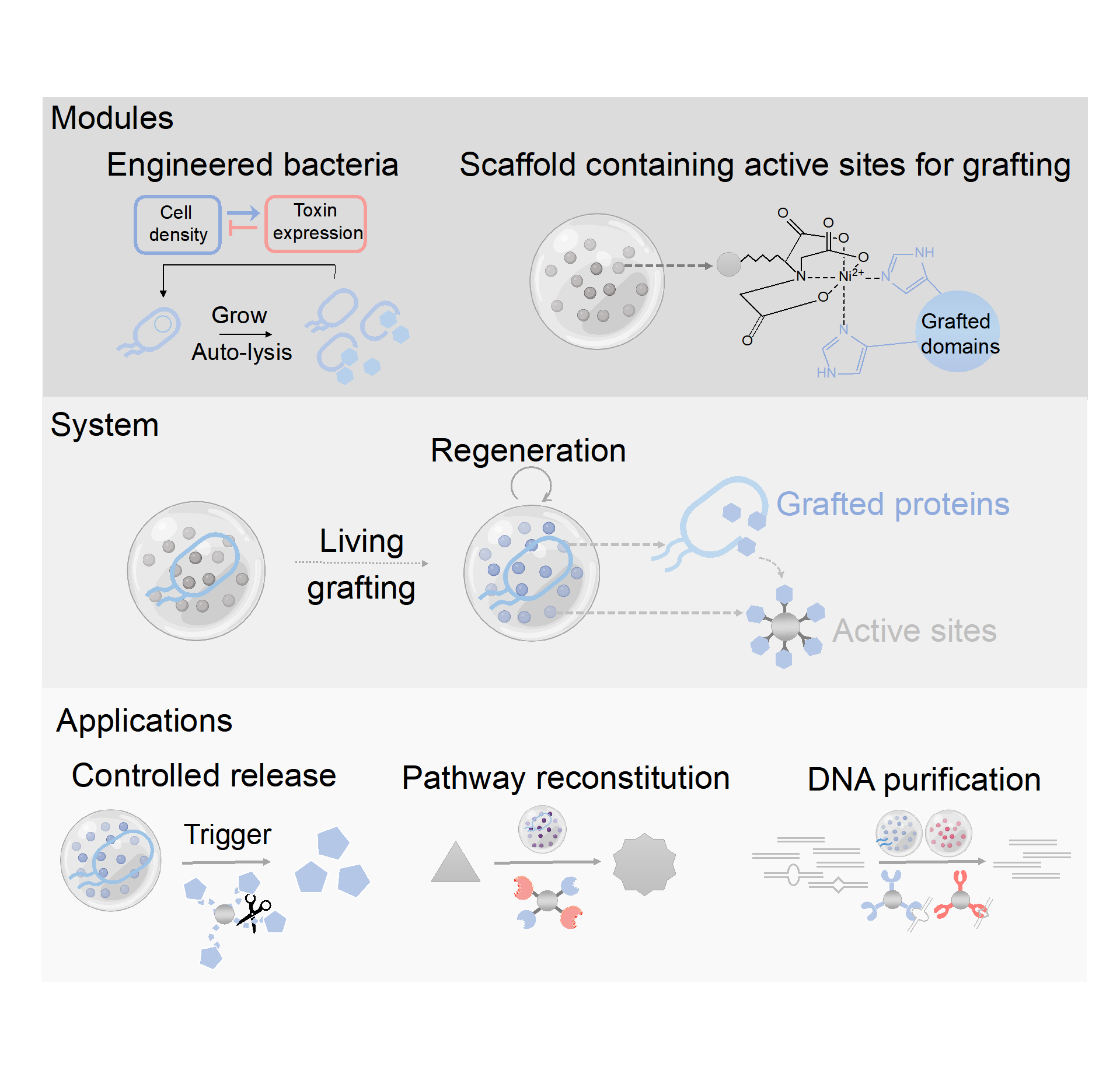
Figure 1. Module, system and downstream applications of MELG (Materials Engineered by Living Grafting)
Living grating materials eliminate mismatched DNA
The MELG system is taken as an example, which recognizes and binds with mismatched DNA to reduce the error rate of synthetic DNA. The rapid progress in synthetic biology has benefited from breakthroughs in DNA synthesis technologies. However, the method of creating nucleotide pools with a chip and assembling double-stranded DNA also accumulated error rates while improving the synthesis efficiency. In order to maintain the integrity and stability of their own genetic material, the organisms in nature have a natural system to fix mismatched DNA. Among them, the DNA binding protein MutS is a key component of the bacterial system for fixing the DNA mismatches. Meanwhile, MutS from different species have different binding specificity for different types of DNA mismatches. The team developed E. coli that could autonomously produce, cleave, and release specific MutS (TaMutS from Thermus aquaticus and TmMutS from Thermotoga maritima), and then generated a MELG system grafted with TaMutS or TmMutS (Figure 2a-b). The experimental data showed that MELG grafted with TmMutS had strong binding ability for GG mismatch, while MELG grafted with TaMutS had higher affinity for AA mismatch (Figure 2c). Therefore, the combination of these two MELG can effectively eliminate the DNA fragments containing AA and GG mismatches (Figure 2d). In addition, the MELG system can be stored for a long time and assembled and used as needed (Figure 2e). The grafted functional proteins can be regenerated, e. g., the team manufactured the E. coli EcMutS grafted with MELG and captured the mismatch DNA marked with the fluorescent signal; the system later underwent elution to remove the grafted EcMutS, but after reproducing the system in the medium, the regenerated MELG could again capture the wrong fragments, demonstrating the regeneration nature of the system (Figure 2f).
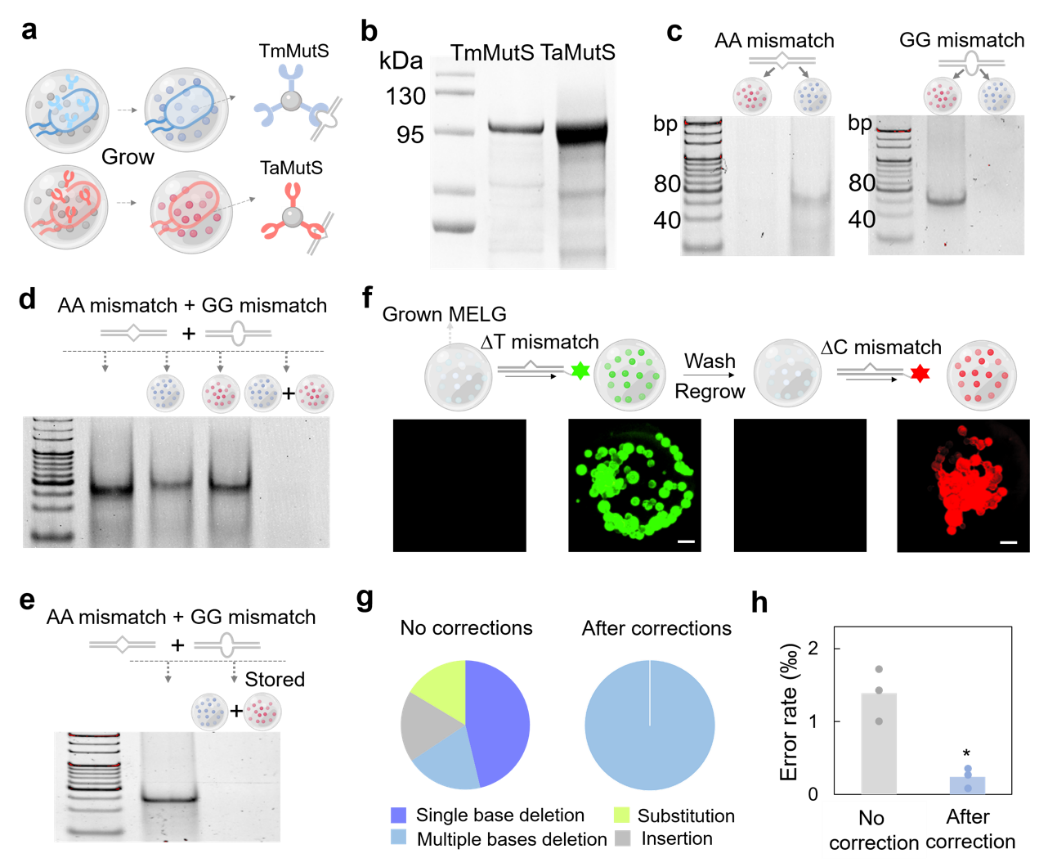
Figure 2. MutS grafting with MELG to remove the mismatched DNA.a. The grafting of different MutS with MELG shows difference in binding with different mismatched DNA. b. E . The coli released TaMutS and TmMutS. c. MutS grafted with MELG has different binding preferences.d. Combined MELG removed most of the mismatched DNA. e. MELG could be used while it was growing.f. MELG was reused after secondary growth.g. Almost all errors were eliminated by MELG except for the multibase deletion. h. The overall error rate decreased after MELG correction.
Finally, the research team synthesized the full-length β -lactamase gene and applied MELG to eliminate accumulated errors during gene synthesis. The results showed that the overall error rate of genes corrected by MELG was reduced from 1.3853 ‰ to 0.2399 ‰, thus improving the overall quality of synthesized DNA (Figure 2g-h). Notably, MELG eliminated all types of errors except multibase deletions, which was consistent with the poor recognition of multibase deletions in MutS (Figure 2g).
Dai Zhuojun, a researcher at Shenzhen Institute of Advanced Technology, Chinese Academy of Sciences, is the corresponding author of the paper. Zhu Runtao, a PhD candidate of National University of Defence Technology (NUDT), is the first author of the paper. Zhang Jiao, a master graduate and now a research assistant of NUDT, has made important contributions to the experimental design, promotion and revision of the article. The research was supported by the National Key Research and Development Program, Guangdong Natural Science Foundation for Outstanding Young Scholars, Shenzhen Institute of Synthetic Biology, etc..
References:
1. Runtao Zhu, Jiao Zhang, Lin Wang, Yunfeng Zhang, Yang Zhao, Ying Han, Jing Sun, Xi Zhang, Ying Dou, Huaxiong Yao, Wei Yan, Xiaozhou Luo, Junbiao Dai, Zhuojun Dai*.Engineering Functional Materials t hrough Bacteria-assisted Living Grafting.Cell systems.2024. DOI: 10.1016/j.cels.2024.02.003