
The widespread application of single-cell technologies generates large amounts of single-cell data, hence increasing the demand for single-cell data analysis and interpretation methods. Although single-cell data can be analyzed with methods such as machine learning, the interpretation of the data is still relatively lacking. Genome-scale metabolic model (GEM) containing a priori knowledge of metabolic networks has become a powerful tool for integrating and interpreting bulk sample omics data. Therefore, GEM may be used to analyze and interpret single-cell data, and the field is indeed on the rise. Integrative analysis of single-cell data using GEM promises to reveal numerous new discoveries in cellular metabolism, explore metabolic differences between cell types and even single cells within complex tissues, and bring new insights into various fields including biotechnology and basic medical research.
Recently, the research team led by Chen Yu with Shenzhen Institute of Advanced Technology, Chinese Academy of Sciences and Eduard J. et al from Charms University of Technology published a review article entitled “Single-cell omics analysis with genome-scale metabolic modeling” in the journal Current Opinion in Biotechnology. This article first reviews the common methods for processing bulk sample omics data with GEM, then introduces the progress in the analysis of single-cell omics data with GEM, and finally points out the existing challenges in the field. Researcher Chen Yu and Dr. Eduard J. Kerkhoven are the corresponding authors of the article, Dr. Johan Gustafsson is the co-first author, and Professor Jens Nielsen and research assistant Yang Jingyu contributed to the writing of the article. This research has been supported by the National Key Research and Development Program, Shenzhen Special Medical Research Program and Shenzhen Science and Technology Program.
Screenshot of the article
Link: https: / / www.sciencedirect.com/science/article/pii/S0958166924000144?via%3Dihub
Given that the methods that analyze bulk sample omics data with GEM are relatively mature, analyzing single-cell data may make direct use of these methods. Therefore, the article first reviews two types of common methods for integrating bulk omics data with GEM. The first method is the model extraction method, where, based on the omics data, the inactive reactions from the reference model (the whole metabolic reaction set including genome annotation) are excluded to generate specificity models, such as tissue or organ specificity models; the second method is to use omics data as a constraint to improve the accuracy of GEM for metabolic flux prediction.
Then, the paper introduces the method of analyzing single-cell data with GEM. Similar to the method of analyzing bulk omics data, it can be roughly divided into two categories, namely, the generation of single-cell specificity models and the prediction of single-cell metabolic flux (Figure 1).
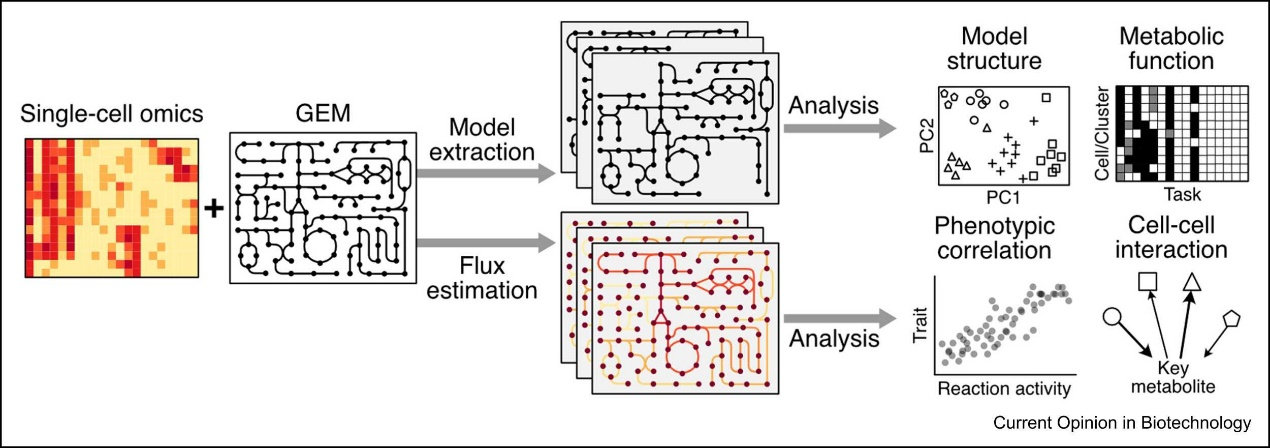
Figure 1. Analysis of single-cell omics data with GEM
Finally, the article summarizes the challenges and prospects in the field. The first challenge comes from the limitations of the single-cell technology itself, which is currently rarely used in the studies of true single cells, so it is difficult to reach the true single-cell level with the analysis integrated with GEM. In the prediction of single-cell metabolic flux, its accuracy is limited by the determination of metabolite exchange at the single-cell level as well as the determination of the single-cell target function. At present, GEM is mainly used to analyze single cell transcriptome data. With the development of single cell proteomics and metabolomics, it is believed that GEM-based multi-omics analysis will be possible in the near future.
About PI and the Research Team
Chen Yu, a researcher, a doctoral supervisor, was selected into the National Major Youth Talent Program (overseas). In 2018, he received his PhD degree in biochemistry from East China University of Science and Technology, and then pursued postdoctoral research with the research team led by Jens Nielsen in Sweden. In 2023, he joined the Institute of Synthetic Biology, Shenzhen Institute of Advanced Technology, Chinese Academy of Sciences. In the past five years, he published articles as first author in academic journals such as PNAS (4), Nature Protocols, Molecular Systems Biology, and Nucleic Acids Research, and published articles as a co-author in journals such as Cell, Nature Catalysis, and Nature Chemical Biology. He has been invited as a reviewer with journals such as PNAS and Nature Communications. At present, he is heading the special medical research project of Shenzhen, and participating in the projects such as National Key Research and Development Program and Shenzhen Science and Technology Program.
The research team led by Chen Yu (https://chenyu.website/) is engaged in systems biology and synthetic biology research, focusing on digital cell modeling based on metabolic network, integrating “dry” and “wet” experimental techniques, and achieving accurate cell simulation to guide rational transformation and artificial synthesis. The research team is now to recruit 2 postdoctoral students with related research backgrounds such as bioengineering, biochemical engineering, fermentation engineering, bioinformatics, computational biology. You are welcomed to email your resume to us at y.chen3@siat.ac.cn.