
Synthetic genomics can reasonably introduce artificial laws through the design and synthesis of the genome to create a synthetic life that can be regulated. In recent years, we have acquired the ability to design and synthesize viral and bacterial genomes, and have successfully constructed minimal prokaryotic genomes. Similarly, a series of advances have been made in the synthesis of eukaryotic genomes, and the first eukaryotic genome synthesis program (Sc2.0 program) has completed the synthesis of all yeast chromosomes. To explore the core functions of the yeast genome, the Sc3.0 program was launched by researcher Dai Junbiao (Shenzhen Institute of Agricultural Genome of Chinese Academy of Agricultural Sciences / Shenzhen Institute of Advanced Technology, Chinese Academy of Sciences), professor Cai Yizhi (University of Manchester) and professor Jef Boeke (New York University) launched Sc3.0 program, with an aim to achieve deep streamlining and reconstruction design of the yeast genomes, and build the first minimal yeast genome. The existence of complex interaction networks between yeast genes poses great challenges to the streamlined design of yeast genomes. The team led by Dai Junbiao established two efficient streamlining strategies for non-repetitive sequences within the scope of the chromosome arms: 1) irrational and random deletion strategy based on the rearrangement system (SCRaMbLE) introduced into Sc2.0 synthetic chromosomes (see the report: Two Articles of BioArt Genome Biology Released | The research team led by Dai Junbiao reports efficient streamlining strategy for artificial genomes - Sc3.0 officially launched https://mp.weixin.qq.com/s?__biz=MzA3MzQyNjY1MQ==&mid=2652521505&idx=5&sn=a23a706d543b96d55c0ef7ead2f97099&scene=21&token=1051366233&lang=zh_CN#wechat_redirect), 2) The synthesis strategy gradually increasing core genes based on a bottom-up rational design principle (Bio ArtMED Nat Commun | Important progress of Sc3.0 : Streamlining and remodeling of chromosome arms in Saccharomyces cerevisiae, https://mp.weixin.qq.com/s?__biz=MzU4OTY0NzE1NQ==&mid=2247567177&idx=3&sn=fe126a7d4bb6da974400c03f1145dff0&scene=21&token=1051366233&lang=zh_CN#wechat_redirect).
However, a large number of repeated sequences still exist in the yeast genome. One of the key regions is the region encoding rDNA, the rDNA region. The rDNA is the core component of the ribosome. To meet the requirement of protein translation for rDNA, eukaryotic cells usually contain hundreds or thousands of copies of rDNA-encoding genes to form highly repetitive rDNA regions. The rDNA regions in eukaryotes usually exist in arrays. For example, the single rDNA array in the S. cerevisiae genome contains repeats of nearly 150 copies, while the rDNA units in the human genome form multiple arrays of tandem repeats on different chromosomes. The highly repetitive arrangement and high transcriptional activity of the rDNA region make it the most unstable region of the genome. All these properties have greatly hindered the manipulation and study of rDNA. How to achieve deep streamlining of highly repetitive and dynamically changing rDNA regions and reduce biological load is an important issue to be solved in building the minimal eukaryotic genome.
On February 6, 2024, the research team led by Dai Junbiao with Shenzhen Institute of Advanced Technology, Chinese Academy of Sciences and the research team led by Cai Yizhi from the University of Manchester published an online research paper entitled “High plasticity of ribosomal DNA organization in budding yeast” in the jouDNAl Cell Reports, which was selected as the cover article of that issue. In this study, CAT, an efficient way to build repeat arrays, was developed. The artificial rearrangement system was introduced into the tandem array of repeated rDNA. It revealed that rDNA sequence with eight copies was sufficient to maintain strain survival. Besides, a yeast model with three arrays was established to explore the influence of the number of rDNA arrays. This study revealed the high plasticity of the organization form of the yeast rDNA region, which was another important achievement of the Sc3.0 program achieved by the team led by Dai Junbiao recently.
Screenshot of the article
Link: https: / / www.cell.com/cell-reports/fulltext/S2211-1247(24)00070-6
Cover: An artificial control system introduced to streamline the rDNA array
To introduce an artificial control system into rDNA regions, researchers have developed CAT, an efficient way to build arrays, which allowed the genome to build synthetic rDNA arrays with a customized sequencing at any location in the genome. With this method, the researchers managed to build ectopic rDNA arrays on chromosome 3 (short) or chromosome 15 (long). Later, with the introduced loxPsym sequence in the synthetic rDNA array and the induced expression of Crerecombinase, the short rDNA arrays on chromosome 3 was deeply streamlined, revealing that rDNA units with eight copies were sufficient to support the survival of S. cerevisiae.
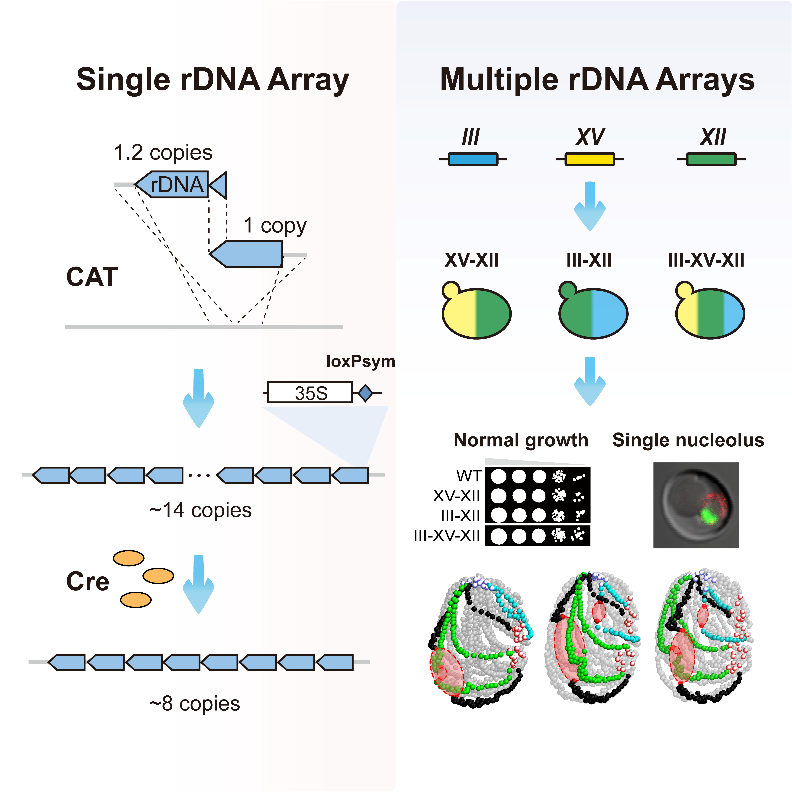
Overall map: construction and streamlining of synthetic rDNA array (left); construction and analysis of multi-array yeast model (right)
The genomes of higher organisms often contain multiple rDNA arrays, distributed across different chromosomes. In order to simulate the tissue form of rDNA in higher organisms and explore the influence of the number of rDNA array, the researchers combined CAT and meiosis and successfully constructed multiple two array strains and three array strains. The results showed that the introduction of extra rDNA arrays significantly changed the 3D conformation of the genome, but had no significant destructive effect on the formation of mono-nucleolus structure, normal growth of strains, and genome expression level, fully demonstrating the inclusiveness of the yeast genome for the organized form of rDNA arrays.
In conclusion, this study achieved efficient customized synthesis of tandem repeated rDNA arrays, and introduced artificial rearrangement system to successfully reduce the rDNA copy number in haploid yeast strains to about 8 copies, which is the lowest rDNA copy number that can support survival. Meanwhile, this study constructed a yeast model of multi-rDNA arrays, analyzed the impact of extra rDNA arrays, and revealed the inclusiveness of yeast cells for multi-array forms. In this study, the concept of synthetic genomics was applied to deeply streamline and synthesize the highly repetitive rDNA arrays, which not only expanded the existing theoretical cognition, but also laid the theoretical foundation for the subsequent construction of the minimal yeast genome and robust simplified yeast chassis, and provided an important research tool for the regulation and analysis of rDNA.
Jiang Shuangying, an associate researcher, Cai Zelin, a research assistant, with Shenzhen Institute of Advanced Technology, Chinese Academy of Sciences, and Wang Yun, an associate researcher with Life Science Institute of Shenzhen BGI Research, and Zeng Cheng, a research assistant with Shenzhen Institute of Advanced Technology, are first authors of the paper. Researcher Dai Junbiao, associate researcher Jiang Shuangying and professor Cai Yizhi are the corresponding authors of the paper. The research was supported by programs such as National Key Research and Development Program, National Natural Science Foundation of China, Guangdong Key Laboratory of Synthetic Genomics, Guangdong Natural Science Foundation, InteDNAtional Great Science Program of Chinese Academy of Sciences, Shenzhen Fund for Outstanding Talents Training, Shenzhen Institute of Synthetic Biology and the Royal Society Newton Senior Scholarship.
For more work about the Laboratory of Dai Junbiao, please visit the link http: / / dailab.org. cn / home. Young talents with keen interest in synthetic biology are welcomed to join us, with liberal wages and benefits available.