
On February 6, 2024, Beijing time, the team led by Luo Xiaozhou and the team led by Jay D. Keasling at Institute of Synthetic Biology, Shenzhen Institute of Advanced Technology, Chinese Academy of Sciences, published an article titled “Pathway evolution through a bottlenecking-debottlenecking strategy and machine learning-aided flux balancing” in the journal Advanced Science.
This study aims to address a key issue in metabolic pathway engineering: genetic epistatic effects have limited its evolutionary potential and adaptability, leading to uncertainty in the evolution. For example, minor modifications of one enzyme may make another a bottleneck in the pathway, which may take thousands of years to enhance or develop new functions. Therefore, how to achieve the effect which may take thousands of years of natural evolution to attain with less evolution time and updates has always been a research difficulty in this field. In view of the above problems, the research team used the automation infrastructure platform technology to determine the controllable evolutionary trajectory, and realized the automatic synchronous evolution of multiple key genes in the metabolic pathway. The study also adopted the ProEnsemble machine learning framework to further mitigate the epistatic effects among genes in the evolutionary pathway, and thus create an efficient generic chassis for the synthesis of flavonoid compounds. This project (DOI:10.1002/advs.202306935) can effectively overcome the technical difficulties of the uncertainty of metabolic pathway evolution. This was another progress in the fields of IT and BT after the UniKP large language model developed by the team led by Luo Xiaozhou for for enzyme mining and evolution in 2023 (Nat.Commun.2023). The study also incorporates the advantages of automation technology and machine learning technologies, which can significantly improve the speed and efficiency of chassis development, reduce research and development time, and reduce the costs. This also provided cutting-edge technology route and new solution for promoting the development of biological intelligent manufacturing.

https://doi.org/10.1002/advs.202306935
(Click “Read the original text” at the end of the article to visit the link)
01 Exploring the “speed mystery” of metabolic pathway evolution: Is there a genetic epistatic effect?
In this study, the team tried to solve a scientific problem in the field of synthetic biology: how to obtain the optimal combination of mutations in a specific evolutionary trajectory. However, different combinations of mutations show different manifestations in different backgrounds, a phenomenon called gene epistatic effect. This effect would lead to uncertainty in pathway evolution, limiting evolutionary potential and adaptability. To verify this phenomenon, the research team took the naringenin metabolic pathway as an example. They first identified TAL missense mutants, then evaluated their adaptability in the context of different gene combinations, and finally confirmed the prevalence of gene epistatic effects in the evolution of the metabolic pathway.
First, the TAL gene was placed in plasmids with different number of copies and weak promoter, while the remaining key metabolic genes stayed in the original plasmid (Figure 1). They revealed that the highest naringenin yield was achieved when the TAL gene was placed in ColE 1, a plasmid with a high number of copies. However, when directly screening the random mutant library of TAL under this background plasmid, it failed to obtain missense mutants with a higher yield, which revealed that complex gene epistatic effects may induce pathway evolution into local optimal solutions. To address this issue, the team placed the random mutant library of TAL in a plasmid with a small number of copies, to create an artificial metabolic bottleneck that ensured that TAL expression and activity was the only factor limiting the yield of naringenin. In this case, the evolutionary trajectory of the mutants is clearer. Before the yield reached the highest yield that could be attained at the plasmid with a large number of copies, theoretically, it would not lead to evolutionary uncertainty due to problems such as toxicity of intermediates or complex regulation. On this basis, the team obtained seven TAL mutants that significantly increased the yield of naringenin under this background and confirmed their mutation sites. Later, the wild-type TAL and seven variants were placed in the plasmids with a medium or large number of copies. It was discovered that the yield of naringenin of all TAL variants was lower than the highest yield of their wild-type variant (357.66 mg / L). The above results confirmed that when the gene TAL was placed in plasmids with a medium or large number of copies (such as ColE1 origin), the gene epistatic effect may mask the missense mutant of naringenin, as a result of which the metabolic pathway often could only reach suboptimal level in direct evolution; this also explained the reason why the pathway evolution often appeared tiny or insignificant.
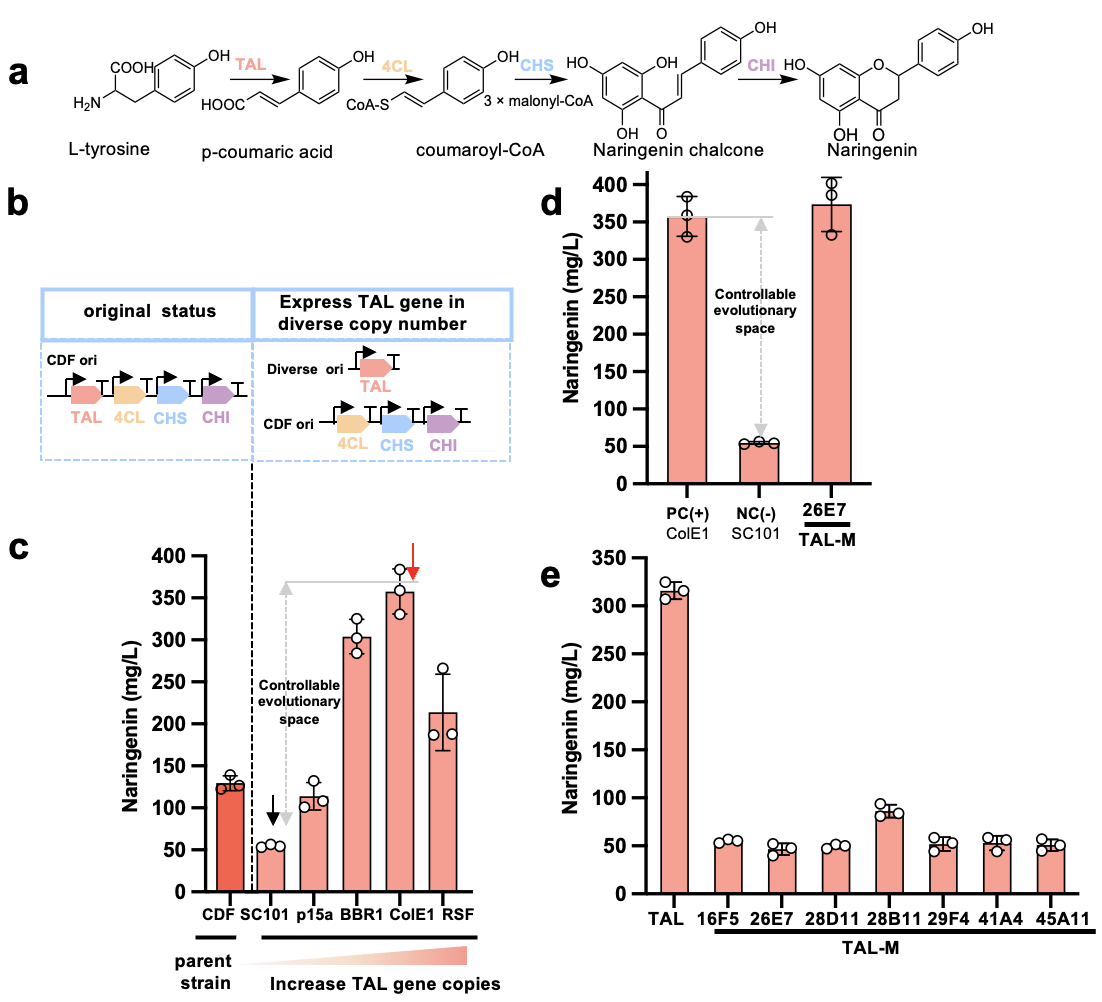
Figure 1 Exploring the epistatic effect of naringenin metabolic pathway genes (taking TAL gene as an example)
02 Automation platform accelerates the synchronous evolution of metabolic pathways: opening up a new vision of enzyme activity and adaptability
By changing the external environment, it can reshape the adaptability of pathway evolution and solve the dilemma that metabolic pathway evolution is stuck in local optimal solution. For this reason, the team lowered the expression level of key genes one by one, changing the evolution adaptability of key genes in metabolic pathways. In order to achieve the synchronous automatic evolution of coenzymes, the research team also adopted the following designs: 1) using the molecular probe system with response to the ultimate product of naringenin as the standard for evaluating the metabolic capacity to achieve a unified screening method; 2) equipped with automatic infrastructure platform technology to achieve the synchronous and iterative evolution of each gene along a clear evolutionary trajectory.
This platform can achieve the routine processes such as bacteria picking, culture, mutation library screening, and alternative mutant product extraction within two weeks. The whole operation is not different from manual operation, which confirms the reliability and accuracy of the platform in the evolution of metabolic pathway. In addition, the platform’s throughput can reach 10,000 clones sorted / time, that is, two genes (5,000 clones / gene /time) or one gene (10,000 clones / gene / time) can evolve at once.
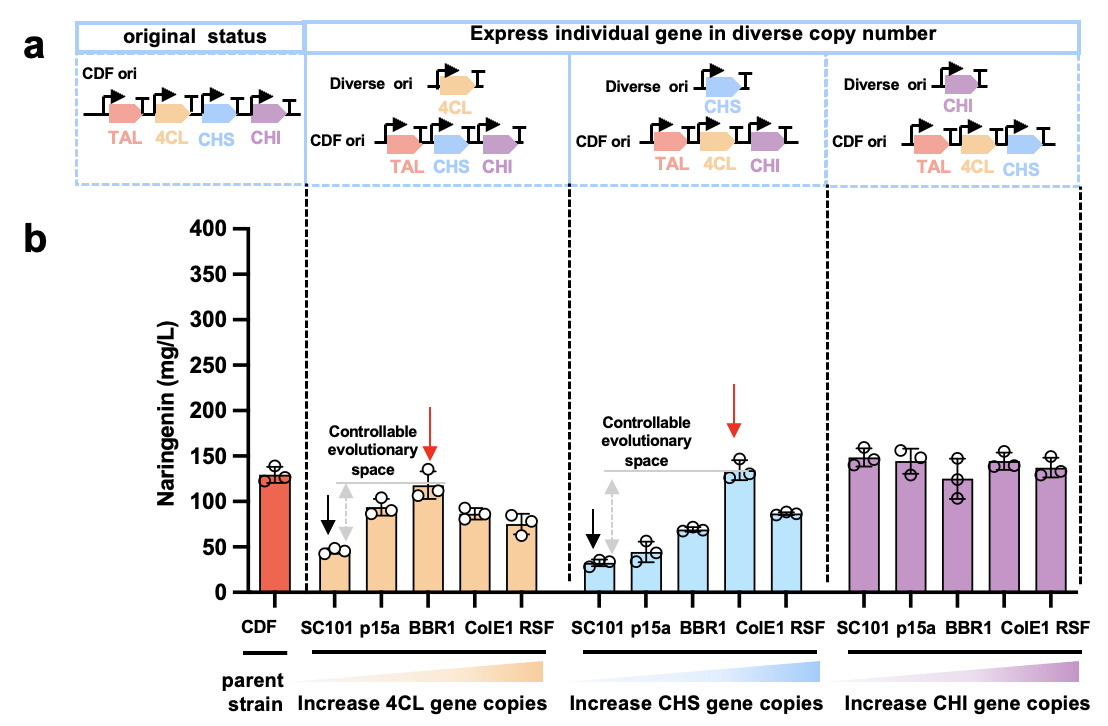
Figure 2 Confirm the evolutionary trajectory of naringenin key genes within a controllable range
Later, the platform technology achieved the directional evolution of 4CL and CHS genes within a clear evolutionary trajectory (Figure 2): the low level expression of each gene (low copy number background) was the starting point of evolution, that is, the artificial bottleneck state; with the increase of copy number, the yield of naringenin reached the highest threshold, that is, the lowest threshold which can be attained by enzyme evolution (artificial de-bottleneck state). Finally, 12 and 57 4CL and CHS mutants were screened from about 5000 clone sublibraries, respectively, and the top 5 and 2 mutants were used for yield and mutation site analysis, respectively. The yields of 4CL-11C1 and CHS-9H9 naringenin were similar to that under the corresponding artificial de-bottleneck state, demonstrating the efficient evolution of metabolic pathways could be achieved through bottleneck-debottleneck strategy within a clear trajectory, and further confirming that epistatic effects may limit the boundaries of pathway evolution. Moreover, the the kcat/KM values of 4CL-11C1 with CHS-9H9 were significantly enhanced by 2.07 times and 4.16 times compared with their wild-type counterparts, respectively (Table 1). Some of the TAL and CHS mutation sites were not in the catalytic core, revealing the potential active distal sites that were difficult to predict could be explored with the aid of the high-throughput platform technology. The discovery overturned the traditional rational design expectations, which also provided us with a new perspective: the aforesaid platform technology can be used to explore the key sites related with enzyme activity or specificity, which were previously unknown. It can help to promote the development of fields such as biological engineering and drug design and provide us with a new key to unlock the potential of enzymes and expand the application scope of biological catalysis.

Figure 3 Parallel evolution of naringenin key genes (automated infrastructure platform) and exploration of intergenic epistatic effects within the scope of a clear evolutionary trajectory
03 Disclosure of genetic epistatic effects: the evolution and adaptability of engineered metabolic pathways
To further decipher whether genetic epistatic effects are prevalent and cause uncertainty in the evolution of metabolic pathways, the team cross-paired the wild-type and beneficial mutants of each gene and assessed the capacity of engineered bacteria in naringenin synthesis (Figures 3 and 4). The results showed a significant reduction in the yield of naringenin in all TAL mutants under the background of combined wild-type genes 4CL and CHS; the yield of naringenin of wild-type TAL increased slightly under the background of combined wild-type mutants 4CL and CHS (Figure 4). Besides, different genetic epistatic effects were also discovered: for example, mutants TAL-26E7 and TAL-28D11 and 4CL-11C1 and CHS-9H9 showed a strong epistatic effect (sign epistasis); the remaining TAL mutants showed positive epistasis; the mutants 4CL-11C1 and TAL-26E7 and CHS-9H9 showed negative epistasis; the mutants CHS-9H9 and TAL-26E7 and 4CL-11C1 showed reciprocal sign epistatic (Figures 3 and 4). These ubiquitous gene epistasis undoubtedly hindered the pathway evolution, causing the pathway evolution to stuck in local optimal solution. Moreover, the task of predicting enzyme mutants with high accuracy was extremely challenging, and the directional evolution mediated by random mutation library was often an event of “luck” or “chance”. Therefore, the simultaneous evolution of each rate-limiting enzyme within a controllable range in a clear trajectory can improve the predictability of the metabolic modification and effectively solve the uncertainty of metabolic evolution.
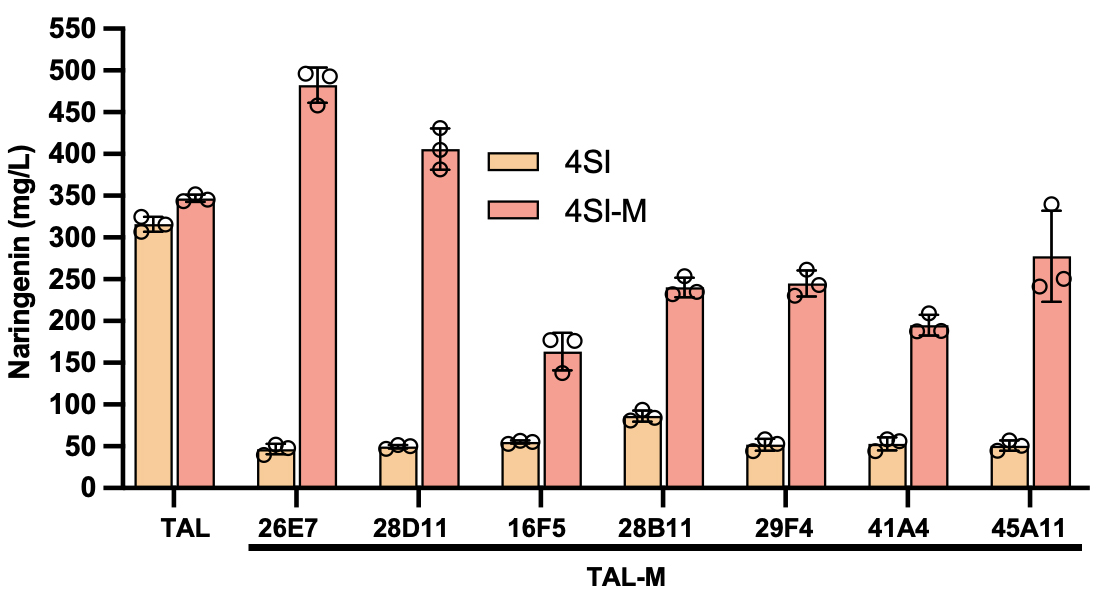
Figure 4 Exploration of the intergenetic epistasis
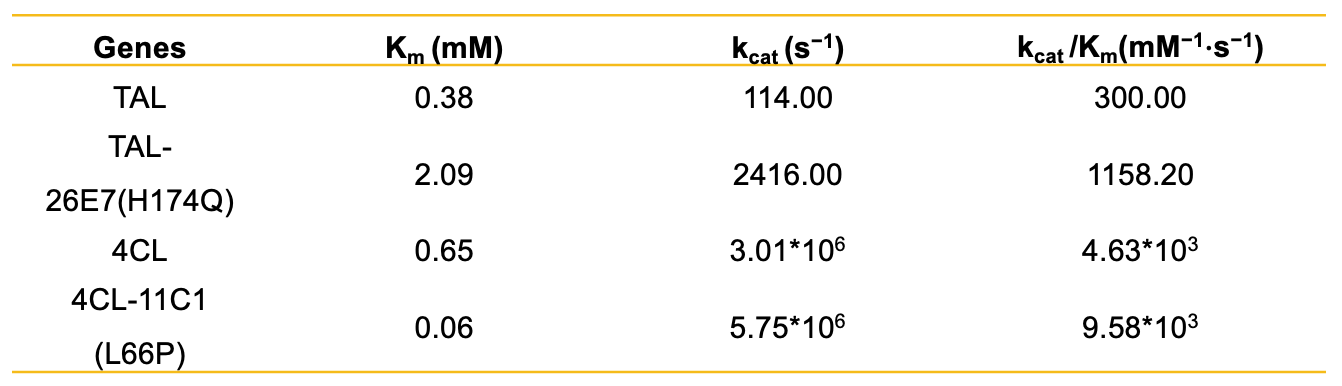
Table 1 Summary of kinetic information of key genes and mutants of naringenin
04 Optimize the combination data of promoter, and IT technology helps to further alleviate the gene epistasis of the evolutionary pathway
Given the influence of gene epistasis, further iterative evolution of the above 3 key genes may induce metabolic pathway imbalance, leading to uncertainty in the evolution. To this end, the team developed the ProEnsemble machine learning framework (Figure 5) to optimize the promoter combination of evolutionary pathways and mitigate the genetic epistasis of evolutionary pathways. In this study, data with different distributions were selected to avoid training from falling into local optimal solution. According to Al3+ signal, a relatively balanced dataset was collected from about 1,000 clones, with the yield of naringenin ranging between 50.8 and 1044 mg/L. The NAR 1.0 strain of Top 1 produced 4.44 times more naringenin than the control group. The Root Mean Square Error (RMSE) of 13 conventional predictors was evaluated by ten-fold cross-validation. Later, the predictors with the least error were integrated successively through forward model selection, and the integration model with the smallest RMSE was selected as the final prediction model. The Pearson Coefficient Correlation (PCC) also reached 0.74, showing a better model correlation between the true and predicted values.
The naringenin yield of Top 5 strains predicted by the ProEnsemble model was higher than 700 mg / L, which was more efficient and accurate than random sampling (5 high-yield strains in 960 samples). However, the dataset still had an unbalanced distribution, which may limit the predictive power of the model, resulting in that the naringenin yield of the Top 5 strain did not surpass that of the NAR 1.0 strain. For this purpose, the training set was further expanded from 1500 clones to optimize the model with datasets above 400 ,500, 600, 700 and 800 mg / L, respectively. Finally, after adding 27 datasets above 600 mg/L in the initial dataset, the model performed best and the PCC increased from 0.74 to 0.82. The results above revealed the importance of the balanced distribution of the dataset in enhancing the model performance. The results showed that all the Top 5 strains predicted in the second round could synthesize naringenin efficiently. NAR 2.0 had the highest yield of 1.21 g / L, 16% higher than that of NAR 1.0 and 5.16 times higher than the initial constructs without promoter optimization. Notably, more than 99.11% of the strains in the random promoter library had a yield below 1g / L, which revealed the possibility that the ProEnsemble integrated model can significantly improve and mine the high-yield strains.
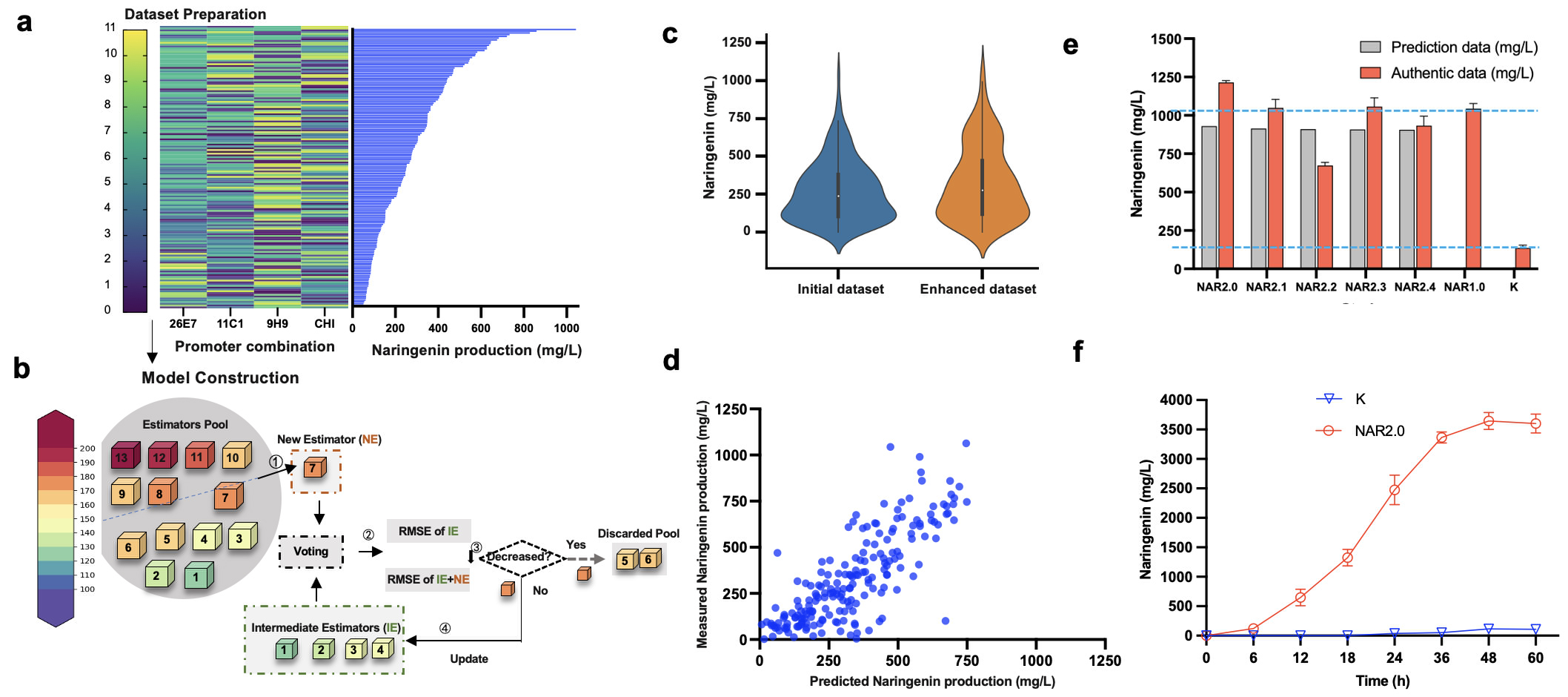
Figure 5 Machine learning framework ProEnsemble further alleviates the intergene epistasis in the evolutionary pathway (machine learning module)
In addition, we performed batch feed fermentation with NAR 2.0 in 1L fermentation bank, where the naringenin yield at 12 h was 660 mg/L, and reached 3.65 g / L at 48h, which was the highest yield of naringenin directly produced from tyrosine as reported in the literature, 3.41 times of the yield with tyrosine as substrate, and 3.02 times that of the coumaric acid intermediate (Figure 5). Given that this study only modified pathway enzymes and promoters, future metabolic engineering strategies could further increase the naringenin yield.
05 Breakthrough in intelligent biomanufacturing: efficient synthesis of flavonoids compounds with universal chassis
Finally, the team achieved efficient synthesis of flavonoids such as genistein, Sakura and hesperetin by overexpressing key synthetic genes, which reached 72.32 mg/L, 223.39 mg/L and 82.50 mg / L. The yield of each flavonoids was higher than the level as reported in the literature (with a series of metabolic engineering modifications) (Figure 6). The above results can reshape the understanding of the synthetic potential of plant flavonoids, provide new ideas and strategies for the production of high value-added compounds, and demonstrate the great potential and application prospect of intelligent biomanufacturing in modern industry.
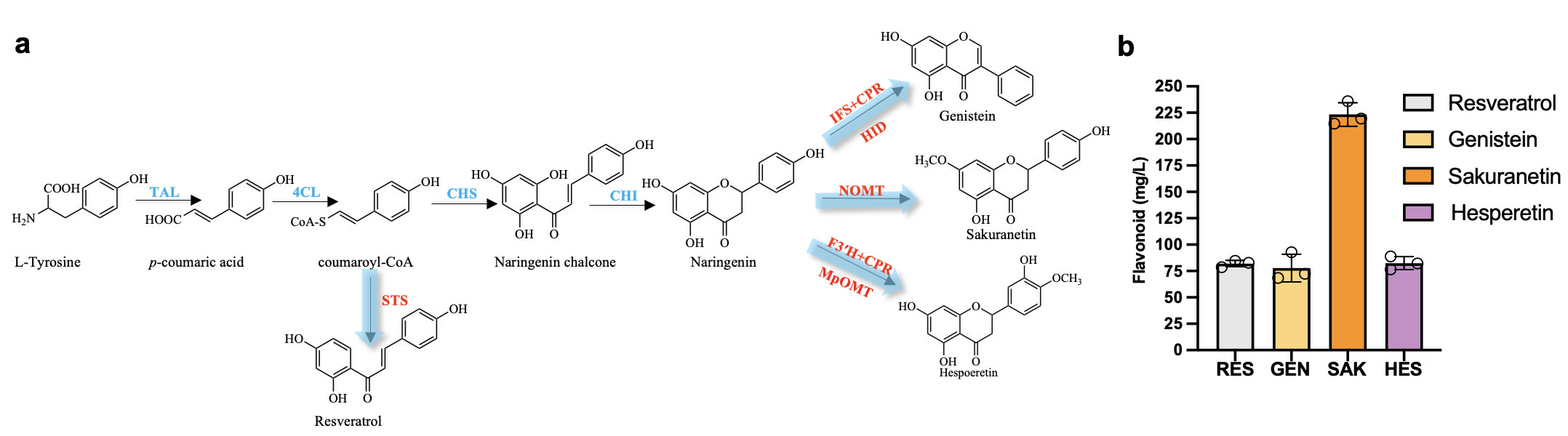
Figure 6 Efficient synthesis of downstream flavonoids by a naringenin chassis
06 Summary and prospect
In view of the universality of gene epistatic effect in pathway evolution and the dilemma of local optimal solution, taking the metabolic pathway of naringenin as an example, based on the automatic infrastructure platform technology, the research team achieved the synchronous evolution of multiple key genes of metabolic pathway within a controllable range of a clear evolutionary trajectory, and with the help of ProEnsemble machine learning framework, further alleviated the gene epistatic effect of evolution pathway, significantly improved the chassis development speed and efficiency, and achieved a leap in the yield of naringenin from laboratory to industrial scale production. This project not only overcomes the technical barriers of the uncertainty of metabolic pathway evolution, reduces the development time and economic cost, but also is of great significance in fields such as metabolic engineering and enzyme engineering and their industrial applications. It provides cutting-edge technical routes and new solutions for intelligent biomanufacturing; and opens up new possibilities for the application of synthetic biology in modern industry.
Luo Xiaozhou, a research fellow and professor Jay D. Keasling with Institute of Synthetic Biology, Shenzhen Institute of Advanced Technology, Chinese Academy of Sciences, are the corresponding author of the article. Deng Huaxiang, an assistant researcher, and Yu Han, a master graduate, are the first authors of the article. Research assistants He Jiahui, Liang Weijie, Deng Yanwu have made important contributions to aspects such as biological experiments. The research was supported by programs such as the National Key Research and Development Program, National Natural Science Foundation, Guangdong Basic and Applied Research Foundation and Shenzhen Science and Technology Program, and platforms such as Shenzhen Key Laboratory of Microbial Drug Intelligent Manufacturing, Shenzhen Institute of Synthetic Biology and Key Laboratory of Quantitative Synthetic Biology. Meanwhile, gratitude was owed to the research assistant Wei Zhenqin for assisting in organizing meetings and discussions and other support work in the project.
Talents Wanted
Luo Xiaozhou is a researcher and doctoral supervisor with the Institute of Synthetic Biology, Shenzhen Institute of Advanced Technology, Executive Director and PI of Synthetic Biochemistry Research Center, deputy chief engineer of Major Science and technological Infrastructure of Synthetic Biology Research in Shenzhen, the founder of Sunrise Biotechnology (Shenzhen) Co., LTD. He received his bachelor’s degree from Nanyang Technological University in Singapore in 2010 and his doctorate degree in chemistry from the Scripps Research Institute in San Diego in 2016 (supervised by academician Peter G. Schultz). During 2016-2019, he was devoted to postdoctoral research at the University of California Berkeley (with academician Jay D Keasling as his partner supervisor). In 2019, he joined Shenzhen Institute of Advanced Technology, Chinese Academy of Sciences. He has been selected as the National Major Talent Project (Youth) Expert, Guangdong Outstanding Youth Scholar, Shenzhen Excellent Youth Scholar, Shenzhen National High-level Talents, Top 10 Outstanding Youth of Nanshan 2023, etc. He has published more than 40 papers in famous academic journals such as Nature, Nature Chemical Biology, Cell Chemical Biology, Nature Synthesis, PNAS and Angewandte Chemie, Advanced Science, Metabolic Engineering. The research team focuses on the research of biochemical processes within living organisms in the field of synthetic biology. Applying various chemical biology methods such as genetic code expansion technology, directional evolution of enzymes, gene mining and metabolic engineering, based on big data machine learning and high throughput automation, the team is engaged in the research on the integrated biosynthesis methods of various natural products and derivatives and applying synthetic biology methods to apply research findings in fields such as pharmaceutical, personalized treatment, and advanced materials.
The research team is now hunting for postdoctoral fellows with an interdisciplinary background in biology, chemistry, bioinformatics, biomedical engineering, or a research background in directional evolution of enzymes, machine learning, high-throughput screening, and biosynthesis of natural and non-natural compounds. You are welcomed to email your resume to us at xz.luo@siat.ac.cn.