
Microorganisms live in changing environments. To survive, bacteria have developed adaptability to these changes. Among them, metabolic flux redistribution and proteome reallocation are common strategies for bacteria to cope with environmental changes[1-2]. For example, when the external nutritional conditions change, the cells will adjust their internal metabolic activities in time to synthesize key metabolites such as amino acid precursors, thus causing the synthesis rate of different types of proteins (such as transporters, ribosomal proteins and metabolic enzymes) in the proteome. We know that the vast majority of chemical reactions in cells are enzymatic reactions. Thus, the reallocation of each protein component in the proteome in turn affects the distribution of flux in the metabolic network. However, existing researches on cellular metabolic fluxes and protein allocation are usually conducted separately. Therefore, we still have fully understood the scientific question of how cells can cross-regulate their metabolic fluxes and protein synthesis to adapt to changing nutrient conditions.
Screenshot of the article
Link: https: / / doi.org/10.1016/j.ymben.2024.01.008
On February 2, Beijing time, the team led by Fu Xiongfei from the Institute of Synthetic Biology, Shenzhen Institute of Advanced Technology, Chinese Academy of Sciences, published their research findings titled “Cross-regulation between proteome reallocation and metabolic flux redistribution governs bacterial growth transition kinetics” on the journal Metabolic Engineering. The team developed a new method called dynamic flow balance analysis (dCAFBA) constrained by protein allocation (Figure 1). This method utilizes the metabolic flow balance method[3] to depict the changes in metabolic state and simulate the changes in protein resource allocation with the aid of a coarse-grained model[4], combining the enzyme constraint on metabolic reaction rate with the regulation of central substrate metabolic rate on protein allocation to establish a comprehensive model for the prediction of bacterial responses to changes in the external environment. This approach is unique in that it can predict the dynamic changes of metabolic flux redistribution in E. coli in the event of changes in nutrient conditions, even without detailed information on enzyme parameters. Therefore, the model has the potential for extensive applications.
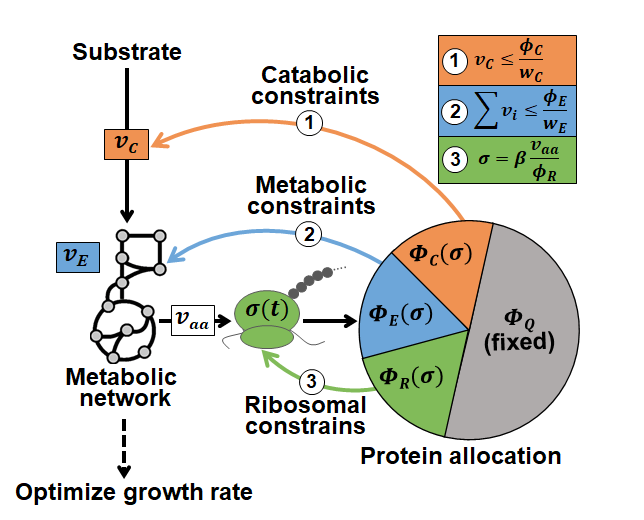
Figure 1 The dCAFBA approach
In their intensive study, the researchers compared the metabolic flux kinetic data predicted by the dCAFBA model with the published proteome data, thus revealing how bacteria adjusted their metabolism and protein synthesis to adapt to changes in nutritional conditions. They discovered that the changes in calculated metabolic flux were consistent with the observed dynamics of the protein level during the nutrient-rich period, and a metabolic bottleneck switch from carbon uptake protein to metabolic enzyme was revealed. This switch disrupted the coordination between metabolic fluxes and protein expression levels, leading to growth overshoot effects that have not been studied before.
In addition, the research team extended the dCAFBA method to study the effect of the heterologous synthesis of valuable compounds on the kinetics of cell growth. Taking lycopene as an example[5], they simulated and predicted changes in cell growth kinetics after exogenous induced expression of the lycopene synthesis gene in E. coli. These findings provide important model tools for designing genetic circuits that regulate the expression of key enzymes in real time as well as increasing the metabolite yield.
This study reveals how microorganisms exquisitely regulate the coupling system of their internal metabolic network and protein synthesis to adapt to changes in environmental conditions. The dCAFBA approach developed in the study not only helps to deepen our understanding of the microbial physiological mechanisms, but also provides theoretical tools for studying the dynamic regulation of metabolic pathways.
Fu Xiongfei, a researcher and Bai Yang, an associate researcher with Shenzhen Institute of Advanced Technology, Chinese Academy of Sciences, are corresponding authors of the article, and assistant researcher Yuan Huili and associate researcher Bai Yang are first authors of the article. Li Xuefei, an associate researcher with the Institute of Synthetic Biology, has made important contributions to the model analysis. The research was supported by the National Key Research and Development Program, the National Natural Science Foundation of China, the Pilot B Special Program of the Chinese Academy of Sciences, the Guangdong Natural Science Foundation, Shenzhen Institute of Synthetic Biology and Shenzhen Major Scientific and Technological Infrastructure for Synthetic Biology Research.
Overview of PI
Fu Xiongfei, a researcher, a doctoral supervisor, Director of Institute of Synthetic Biology, Shenzhen Institute of Advanced Technology, Chinese Academy of Sciences, Deputy Director of Key Laboratory of Quantitative Synthetic Biology (Chinese Academy of Sciences), Vice President of Shenzhen Institute of Synthetic Biology.
The main research interests of the research team: through the combination of quantitative theory and synthetic reconstruction, the interaction between intracellular gene circuit and chassis, and the design principles of orderly structure design of spatial expansion biological systems.
The research team is now recruiting: postdoctoral fellows, research assistants, technicians and visiting students with related research backgrounds such as mathematics, physics, biology, and complex systems. You are welcomed to to contact us at Yang.bai@siat.ac. cn, with your resume and email title indicating “Position to apply for-university name-major-your name”.
Homepage of the Laboratory: http://isynbio.siat.ac.cn/fulab
References:
1. Jens Nilsen (2003) It is all about metabolic fluxes.Journal of Bacteriology 185, 7031-7035.
2. You CH , et al.(2013) Coordination of bacterial proteome with metabolism by cyclic AMP
signalling.Nature 500(7462):301-306.
3. Orth, J.D., Thiele, I.& Palsson, B.O.What is flux balance analysis?Nature Biotechnology 28,
245-248 (2010).
4. Erickson DW , et al.(2017) A global resource allocation strategy governs growth transition
kinetics of Escherichia coli.Nature 551(7678):119-+.
5. Choi, H.S., et al., In Silico Identification of Gene Amplification Targets for Improvement of Lycopene Production.Applied and Environmental Microbiology, 2010. 76(10): p.3097-3105.