
Genome-scale metabolic model (GEM) is a mathematical model to systematically describe cellular metabolism. The “dry” experiments through computer modeling can guide and partially replace “wet” experiments which may be time-consuming and laborious or even cannot be realized, which can greatly improve research efficiency. Thus far, researchers have constructed GEM of thousands of species, and these models are widely used in areas such as biomanufacturing and life health, especially playing an important role in such aspects as cell factory design and human disease research.
The current weakness of GEM lies in the lack of cell physiological information constraints, so researchers have long been committed to integrating various kinds of information in GEM to improve the model. Among them, the model integrating enzyme information, namely the Enzyme-constrained model (ecModel), has better prediction performance and broader application prospect compared with the classical GEM. However, on the one hand, it requires a large number of enzyme parameters to integrate the enzyme constraints, and the lack of parameters hinders the popularization of the ecModel. On the other hand, the threshold is high for construction and use of the ecModel, which will hinder the promotion of the model.
On January 18, 2024, Beijing time, Chen Yu from Shenzhen Institute of Advanced Technology, Chinese Academy of Sciences and Eduard J. Kerkhoven et al from Charms University of Technology, jointly published an article entitled “Reconstruction, simulation and analysis of enzyme-constrained metabolic models using GECKO Toolbox 3.0” in the journal Nature Protocols. This article introduced the latest version of the toolbox GECKO 3.0 for enzyme constraint model development and application, optimized the old version of [1,2] and introduced the enzyme parameter prediction [3] and selection modules, making it possible to construct the ecModel for any species. In addition, this article, as a guide to GECKO 3.0, describes in detail the construction, simulation and analysis of the model, which can greatly reduce the threshold of model. The first authors of the article are Chen Yu, Johan Gustafsson and Albert Tafur Rangel, and the corresponding author is Eduard J.Kerkhoven.

Screenshot of the article
Link: https: / / www.nature.com/articles/s41596-023-00931-7
First, the article introduces the principle of the model (Figure 1) and the new functions of GECKO 3.0, such as introducing the modules for enzyme parameter prediction and selection and the option for building a light version of the model (light ecModel).
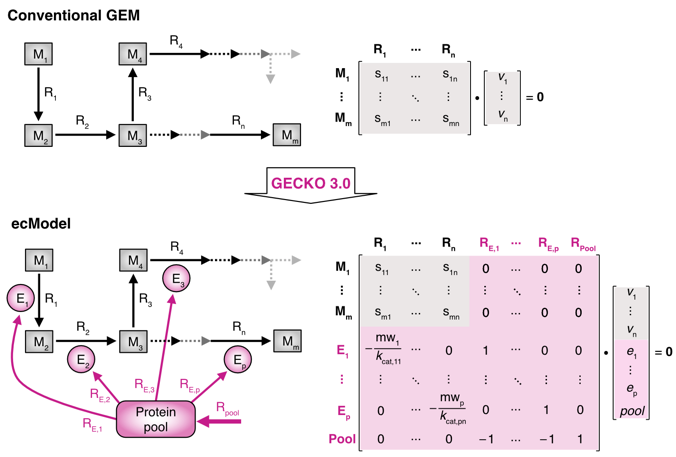
Figure 1. Principle of the ecModel
Later, the article provides a detailed tutorial on the use of GECKO 3.0, including the documentation and data preparation stage (Stage 0), the model building stage (Stages 14, see Figure 2), and the model simulation and analysis stage (Stage 5), involving a total of 79 steps. In addition, the construction and application of ecModel for S. cerevisiae Saccharomyces cerevisiae and human Homo sapiens are taken as an example to explain each step in detail.
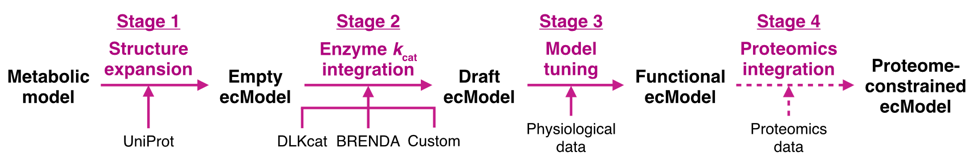
Figure 2. The steps for building an ecModel
Finally, the anticipated results and troubleshooting parts of the article can help the users in self-inspection and debugging. In addition, the code repository released with the article (https: / / github.com / SysBioChalmers / GECKO /) provides an online communication platform to ensure efficient communication between users and developers.
In summary, this article facilitates the construction and application of ecModel, which can promote the popularization of the next-generation metabolic models.
About PI and the Research Team
Chen Yu, a researcher, a doctoral supervisor, was selected into the National Major Youth Talent Program (overseas). In 2018, he received his PhD degree in biochemistry from East China University of Science and Technology, and then pursued postdoctoral research with the research team led by Jens Nielsen in Sweden. In 2023, he joined the Institute of Synthetic Biology, Shenzhen Institute of Advanced Technology, Chinese Academy of Sciences. In the past five years, he published articles as first author in academic journals such as PNAS (4), Nature Protocols, Molecular Systems Biology, and Nucleic Acids Research, and published articles as a co-author in journals such as Cell, Nature Catalysis, and Nature Chemical Biology. He has been invited as a reviewer with journals such as PNAS and Nature Communications. At present, he is heading the special medical research project of Shenzhen, and participating in the projects such as National Key Research and Development Program and Shenzhen Science and Technology Program.
The research team led by Chen Yu (https://chenyu.website/) is engaged in systems biology and synthetic biology research, focusing on digital cell modeling based on metabolic network, integrating “dry” and “wet” experimental techniques, and achieving accurate cell simulation to guide rational transformation and artificial synthesis. The research team is now to recruit 2-3 postdoctoral students and research assistants each with related research backgrounds such as biological engineering, biochemical engineering, fermentation engineering, bioinformatics, computational biology. You are welcomed to email your resume to us at y.chen3@siat.ac.cn.
References
[1] Sánchez BJ, et al.Molecular Systems Biology, 2017, 13(8): 935.
[2] Domenzain I, et al.Nature Communications, 2022, 13(1): 3766.
[3] Li F, et al.Nature Catalysis, 2022, 5(8): 662-672.