 You are here:
Home
>>
RESEARCH
You are here:
Home
>>
RESEARCH
Research
-
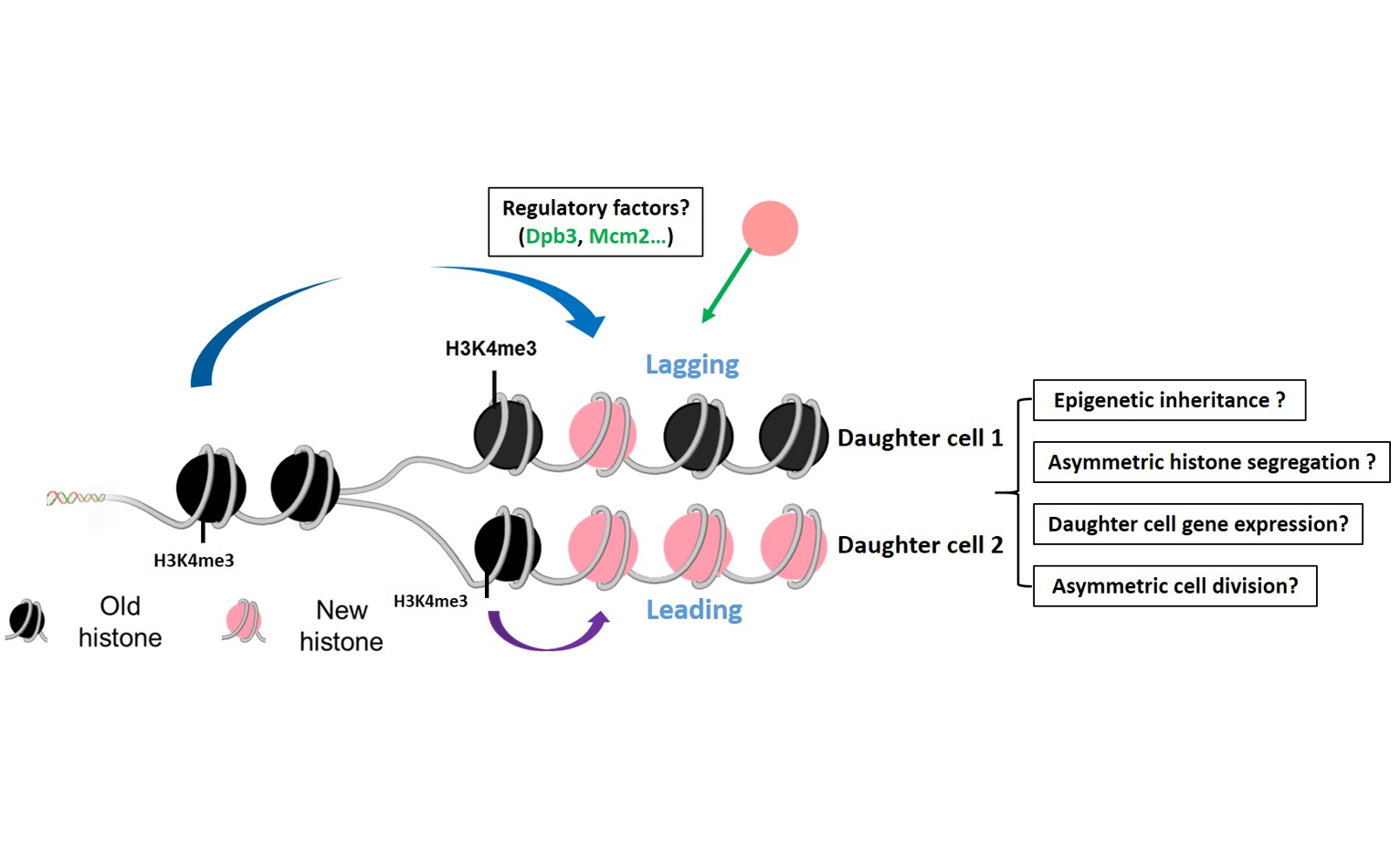
Epigenetic inheritance mechanism
Chromatin states encode epigenetic information and must be stably transmitted to daughter cells to maintain gene expression and cell identity. However, the molecular mechanism of how the parental histone (the epigenetic information carrier) transferred to replicating DNA and how the marks on parental H3-H4 are copied to new (H3-H4)2 tetramers is largely unknown, which hinders our understanding of transmission of epigenetic information into daughter cells. We will study molecular the mechanism epigenetic inheritance in both yeast and mammalian cells, and the consequence of the epigenetic inheritance regulation.
-
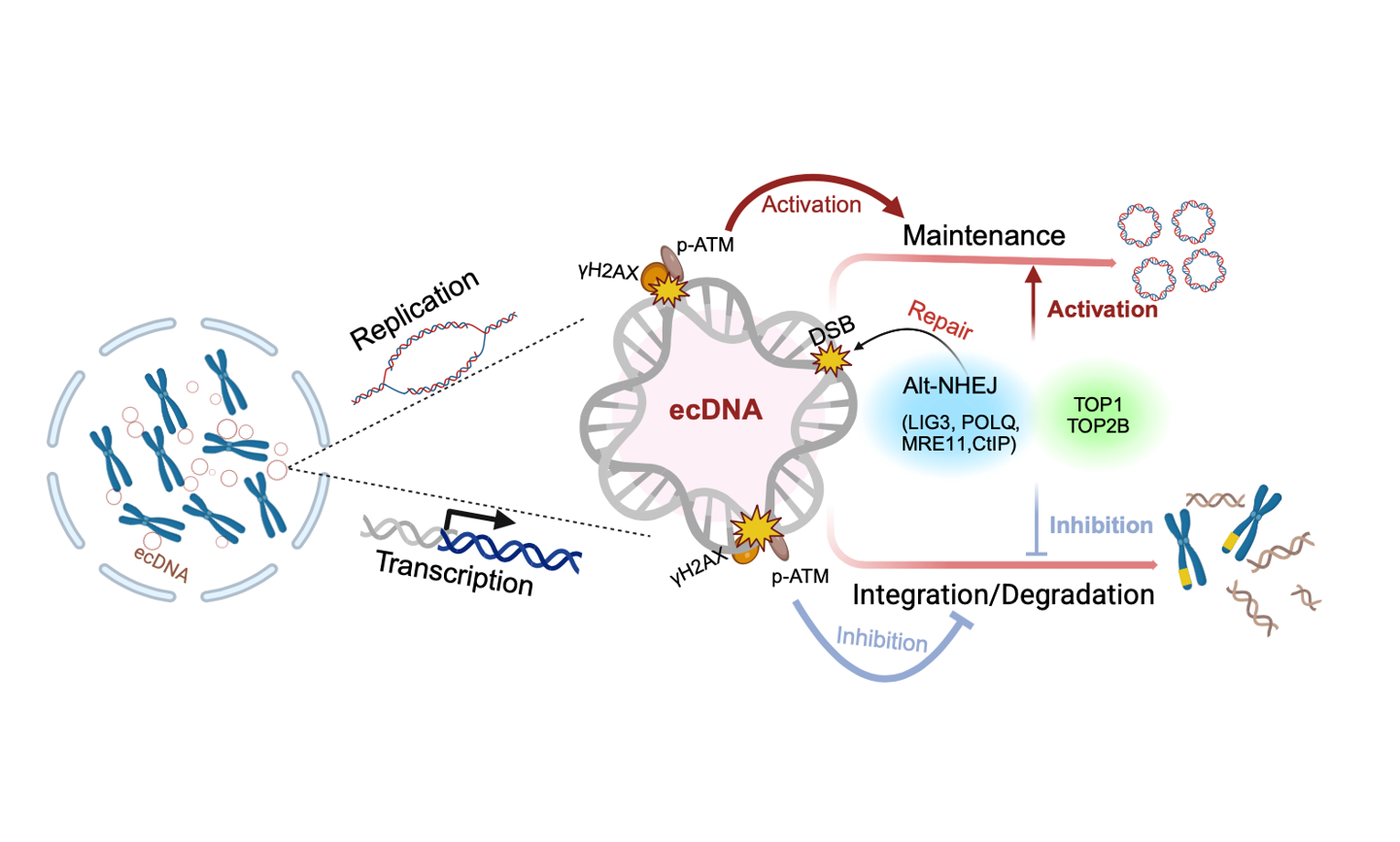
Extrachromosomal DNA (ecDNA)
Extrachromosomal DNA (ecDNA) drives the evolution of cancer cells. However, the functional significance of ecDNA and the molecular components involved in its replication and maintenance remain largely unknown. Here, using CRISPR-C technology, we generated ecDNA-carrying (ecDNA+) cell models. By leveraging these models alongside other well-established systems, we demonstrated that ecDNA can replicate and be maintained in ecDNA+ cells. The replication of ecDNA activates the ATM-mediated DNA damage response (DDR) pathway. Topoisomerases, such as TOP1 and TOP2B, play a role in ecDNA replication-induced DNA double strand breaks (DSBs). A subset of these elevated DSBs persists into the mitotic phase and is primarily repaired by the alternative non-homologous end joining pathway (alt-NHEJ), which involves POLθ and LIG3. Correspondingly, ecDNA maintenance requires DDR, and inhibiting DDR impairs the circularization of ecDNA.
-
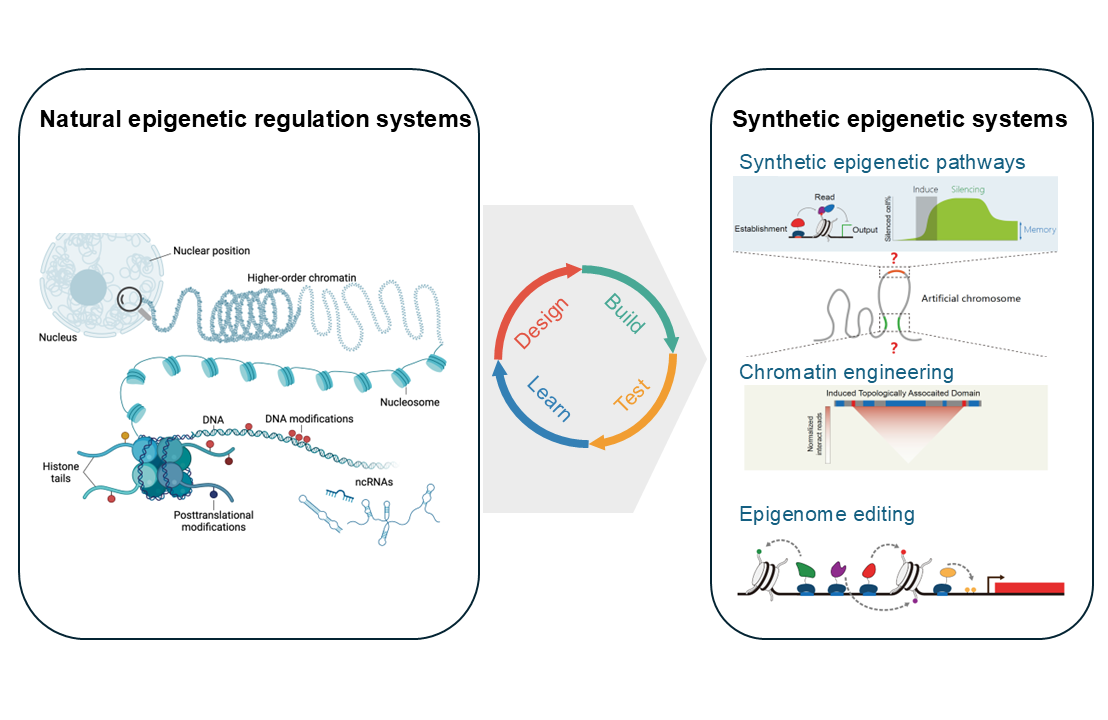
Synthetic Epigenetic
Unlocking the Epigenetic Code: Synthetic Biology Meets Chromatin Engineering. Emerging insights into epigenetic regulation have revealed chromatin-associated pathways as master conductors of cellular identity, aging, and specialization. Our laboratory sits at the cutting edge of this revolution, uniquely bridging synthetic biology with epigenetic engineering to reimagine how we understand and manipulate life’s molecular control systems. We pursue three transformative research frontiers: 1. Designing Epigenetic Control Modules We hunt for nature’s hidden toolbox – discovering novel chromatin regulators (including DNA/histone modifiers and epigenetic “switches”) – then rewire them into synthetic circuits. Like building molecular dimmer switches for genes, we engineer these components to create programmable systems that precisely tune chromatin states, opening new possibilities for controlling cellular behavior. 2. Building Smart Cell Factories Imagine microbes or mammalian cells with upgradable “epigenetic software”! We redesign chromatin architecture in industrial cells using synthetic pathways, achieving two breakthroughs: Supercharged Bioproduction: Lock cells into high-performance metabolic states for sustainable chemical synthesis Living Epigenetic Labs: Create cellular models to crack unanswered questions in development and disease 3. Precision Editing of Cellular Memory We pioneer next-generation tools that edit epigenetic marks without altering DNA. Our dCas9-based editors act like molecular erasers and pens for chromatin, currently being optimized to: Reset cancer cells by wiping out harmful epigenetic “memories” driving tumor growth Develop safer epigenetic therapies that preserve genome integrity
Grants





