
On March 16, 2024, the research team led by Dai Lei with the Synthetic Microbiome Research Center of Shenzhen Institute of Advanced Technology, Chinese Academy of Sciences and Shenzhen Institute of Synthetic Biology published their latest research findings on quantitative ecology of gut microbiome in Nature Communications, a sub-journal of Nature, with the title of “Data-driven prediction of colonization outcomes for complex microbiota”. The team proposed and verified a data-driven research approach for the microbiota, using the species composition of microbiota to accurately predict the colonization outcomes of exogenous species. Dr. Wu Lu with Shenzhen Institute of Advanced Technology and Dr. Wang Xuwen from Harvard Medical School are the co-first authors. Researcher Dai Lei and associate professor Liu Yangyu of Harvard Medical School are the corresponding authors of the article.
Link of the article: https: / / www.nature.com/articles/s41467-024-46766-y
The human gut microbiome plays an important role in resisting the colonization of pathogenic bacteria. This phenomenon is called colonization resistance and refers to the resistance of the microbiota to exogenous species. It was revealed that both the species composition and colonization resistance of the gut microbiota showed significant individual differences. It is very difficult to comprehensively analyze and interpret the interaction network between species for the gut microbiome of a specific individual. Is it possible to accurately predict the colonization outcomes of exogenous species? Is the colonization resistance determined by the key species? In this study, the research team proposed and verified that data-driven research paradigms can be used to predict and understand the functions of complex microbiota, which in turn can achieve accurate regulation of community functions by introducing key species.
First of all, through the analytical derivation and calculation simulation with Lotka-Volterra model in a broad sense, the team proved that when the training data amount was equivalent to species diversity of microbiota, machine learning models such as random forest and neural ODE can be applied to accurately predict the colonization outcomes and stable abundance of exogenous species based on the species composition of gut microbiota (Figure 1).

Figure 1. By sampling microbiota of different species composition as training data, machine learning models can be used to accurately predict the colonization outcomes of exogenous species
Then, in the in vitro culture system of the human gut microbiota, the team conducted large-scale colonization experiments in about 300 human gut microbiota composed of about 300 species, with pathogenic bacteria Enterococcus faecium and probiotics Akkermansia muciniphila as representative species (Figure 2). The experimental results revealed that the colonization outcomes of exogenous species varied significantly between individuals. Stable abundance may also differ by more than two orders of magnitude in communities that can successfully colonize. Moreover, the species diversity and colonization resistance of the gut microbiota were significantly reduced after antibiotic treatment, which was consistent with previous empirical observations and theoretical studies. For colonization experimental datasets of two different species, machine learning models can be used to accurately predict colonization outcomes based on the initial species composition of the gut microbiota.
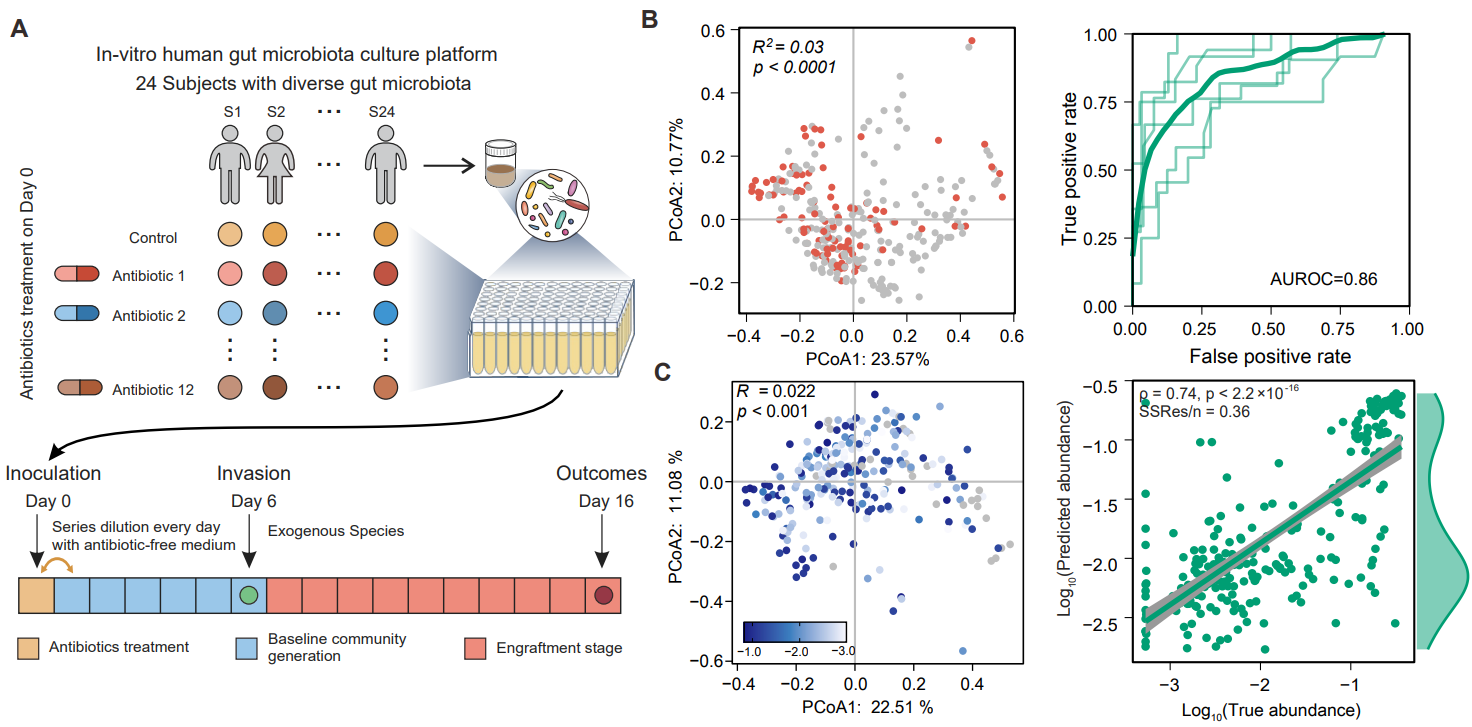
Figure 2. For the gut microflora from different individuals, colonization experiments with exogenous species are conducted and the experimental results are predicted
Finally, the team conducted hypothetical experiments removing specific species using machine learning models to further understand the ecological mechanisms of colonization resistance (Figure 3). The model inferred that the majority of symbiont were weakly antagonistic to exogenous species, which was consistent with the emergent characteristics of colonization resistance in complex communities. Moreover, it was discovered that the presence of certain key species could significantly improve the colonization resistance. For example, Enterococcus faecalis has a very significant inhibitory effect on the colonization of Enterococcus faecium, which was verified by experiments. This result suggests that data-driven research methods can be used to infer key species of colonization resistance, which in turn can guide the accurate regulation of the microbiome. By regulating the composition of microbiota, it can prevent the colonization of pathogenic bacteria or promote the colonization of probiotics, which has important applications in human health and agricultural production.
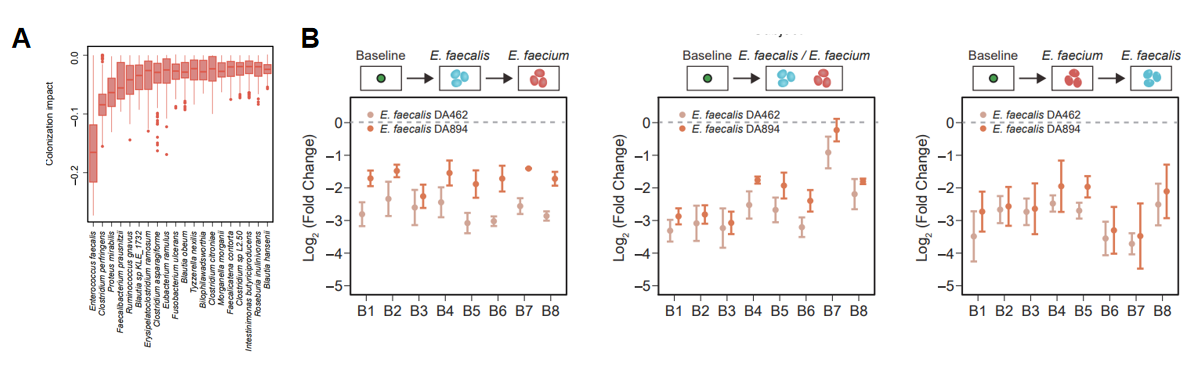
Figure 3. Inference and verification of the key species that determine colonization resistance of gut microbiota
Special acknowledgments are given to the major scientific and technological infrastructure of synthetic biology research in Shenzhen, whose automatic DNA extraction platform ensures the accuracy and reproducibility of the experiment. This research was funded by the National Key Research and Development Program (No.2019YFA0906700), the National Natural Science Foundation of China (No.31971513, No.32100089), Guangdong Provincial Natural Science Foundation (No.2022A1515011513), Shenzhen Key Laboratory of Intelligent Manufacturing of Microbial Drugs (ZDSYS20210623091810032) and Shenzhen Institute of Synthetic Biology.