Jing wang 1 2 †, Wenbing Cao 3 †, Zhaohui Zheng 3 †, Yan He 1, Jiliang Deng 1, Xiaocong Lu 1, Junfeng Shen 3, Xiaoyu Yang 4, Yanhui Liu 5, Shi Xiao 1, Tao Yu 3, Hongting Tang 1.Harnessing the yeast nucleus for the efficient production of valuable monoterpenes.Metabolic Engineering,2026,1096-7176.
Linhai Xie a,b,1 , Shuo Tian a,b,1 , Zhehao Jin b , Tiantian Zou b , Mingxin Chang b,c , Hongting Tang d , Tao Yu b,* , Zhoukang Zhuang b,*.Systematic engineering of cell wall for improving single cell protein(SCP) production.Journal of Biotechnology,2026,173-183.
Tiantian Zou,∥ Xuemei Zhao,∥ Jike Xue, Soo-Un Kim, Tao Yu,*and Zhehao Jin*.High-Level Ferulic Acid Production in Engineered Yeast Enables Discovery and Biosynthetic Application of Novel Enzymes from Piper nigrum.Journal of Agricultural and Food Chemistry,2026,3752-3766.
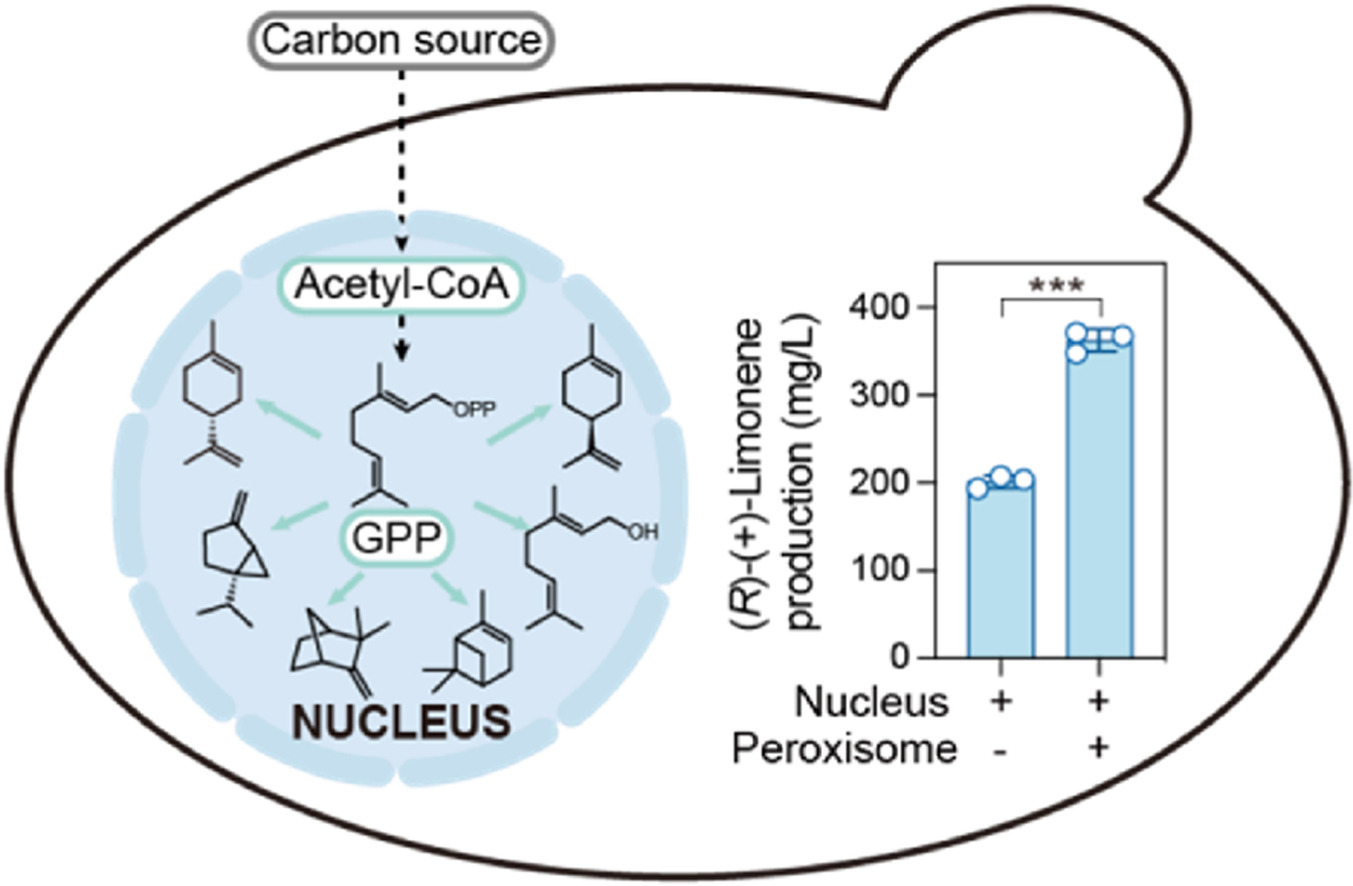
Lon JR, Zhao X, Mamatrixat G, Zhuang Z, Jin Z, Yu T*, Wang J*, Tang H*. Peroxisomal Compartmentalization of the Methylerythritol-4-phosphate Pathway Alleviates Cellular Stress and Enhances Geraniol Production in Saccharomyces cerevisiae. ACS Synthetic Biology, 2025.
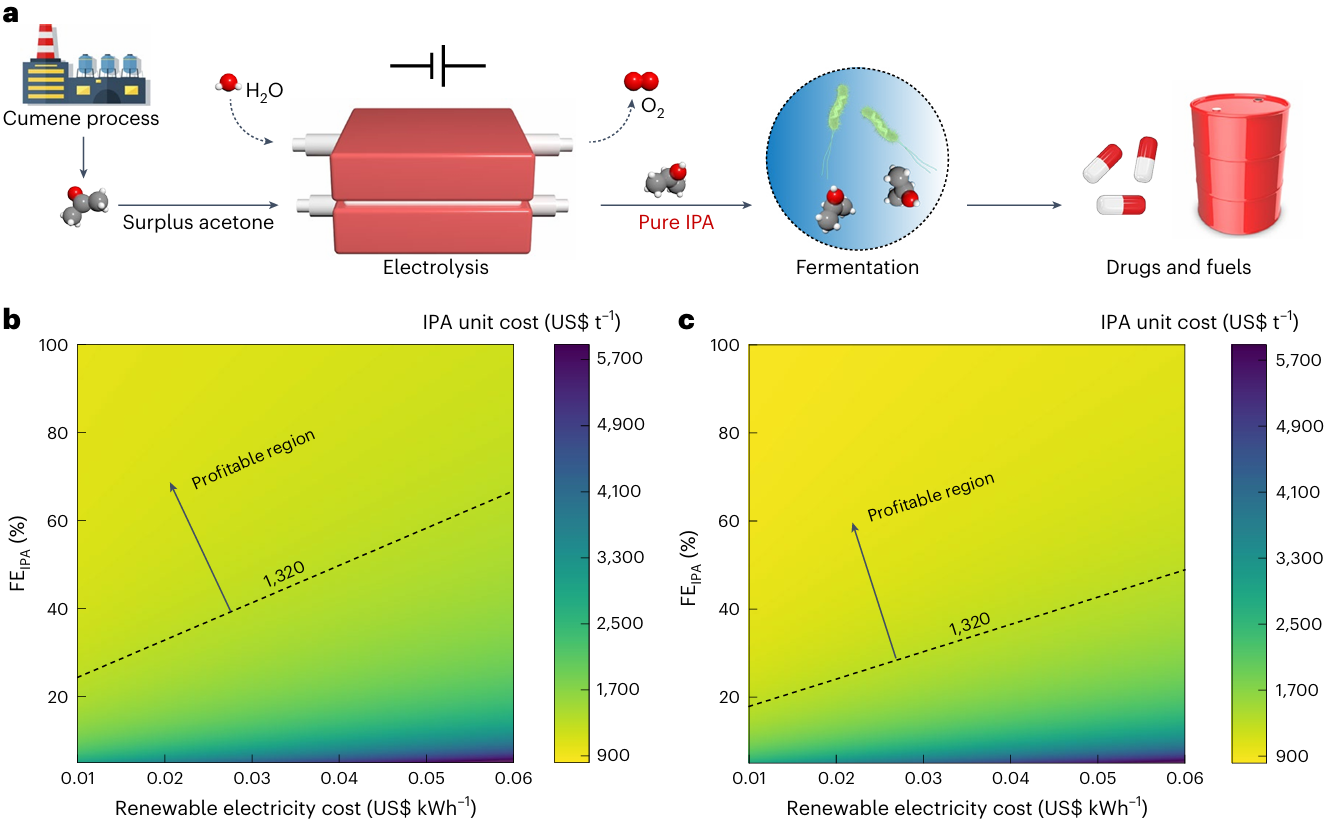
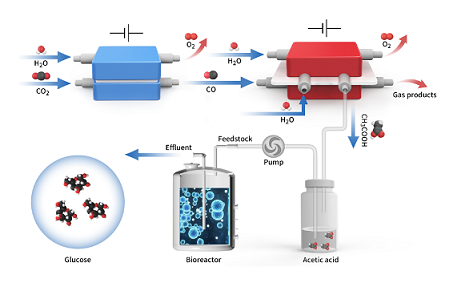
Liu C#, Zhao J#, Tang H#, Xue J, Xue W, Li X, Li H, Jiang Q, Zheng T*, Yu T*, Zeng J*, Xia C*. Upcycling surplus acetone into long-chain chemicals using a tandem electro-biosystem. Nature Sustainability, 2025, 8: 806-817.
Li Y, Wang X, Chen K, Zhuang Z, Tang H, Yu T*, Cao W*. Efficient production of 2′-fucosyllactose in Pichia pastoris through metabolic engineering and constructing an orthogonal energy supply system. Synthetic and Systems Biotechnology, 2025, 10(3):807-815.
Lu X#, Chang M#, Li X#, Cao W, Zhuang Z, Wu Q, Yu T*, Yu A* and Tang H*. Metabolic engineering for sustainable xylitol production from diverse carbon sources in Pichia pastoris. Microbial Cell Factories, 2025, 24: 59.
Chen K#, Maimaitirexiati G#, Zhang Q, Li Y, Liu X, Tang H, Gao X, Wang B*, Yu T*, Guo S*. CRISPR-Cas9-based one-step multiplexed genome editing through optimizing guide RNA processing strategies in Pichia pastoris. Synthetic and Systems Biotechnology, 2025, 10(2): 484-494.
Yang Y, Liu Q, Tian S, Yu T. Research progress in energy metabolism design of cell factories. Chinese Journal of Biotechnology, 2025, 41(3): 1098-1111.
Sun D, Wu L, Lu X, Li C, Xu L, Li H, He D, Yu A, Yu T, Zhao J, Tang H, Bao X. Engineering transcriptional regulatory networks for improving second-generation fuel ethanol production in Saccharomyces cerevisiae. Synthetic and Systems Biotechnology, 2024, 10(1): 207-217.
Zhuang Z#, Wan G#, Lu X, Xie L, Yu T* and Tang H*. Metabolic engineering for single-cell protein production from renewable feedstocks and its applications. Advanced Biotechnology, 2024, 2:35.
Bai Z, Guo S, Yang Y, Zhuang Z, Cao W, Yang Y, Yu T, Tang H. Microbial production of food compounds with carbon dioxide and derived low-carbon molecules as substrates. Chinese Journal of Biotechnology, 2024, 40(8):2731-2746.
Zeng T, Jin Z, Zheng S, Yu T, Wu R. Developing BioNavi for Hybrid Retrosynthesis Planning. JACS Au, 2024, 4(7):2492-2502.
Guo S,Zhang Q,Gulikezi·M,YANG Y,Yu T*. Advances in microbial production of liquid biofuels. Synthetic Biology Journal, 2024, 5:1-16.
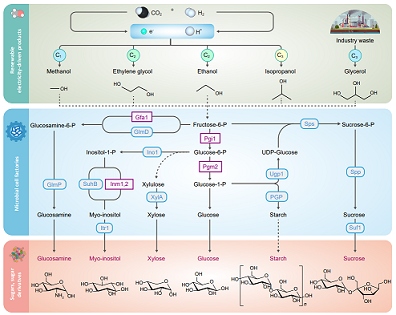
Tang H#, Wu L#, Guo S#, Cao W, Ma W, Wang X, Shen J, Wang M, Zhang Q, Huang M, Luo X, Zeng J, Keasling J*, Yu T*. Metabolic engineering of carbohydrate-derived foods and chemicals production in yeast. Nature Catalysis, 2024, 7:21-34.
Wang X, Li Y, Jin Z, Liu , Gao X, Guo S*, Yu T*. A novel CRISPR/Cas9 system with high genomic editing efficiency and recyclable auxotrophic selective marker for multiple-step metabolic rewriting in Pichia pastoris. Synthetic and Systems Biotechnology, 2023, 8(3):445-451.
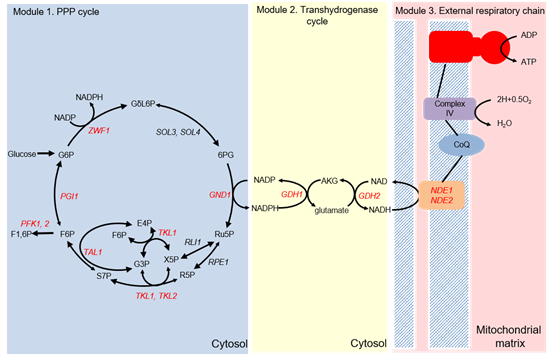
Yu T*, Liu Q, Wang X, Liu X, Chen Y and Nielsen J*. Metabolic reconfiguration enables synthetic reductive metabolism in yeast. Nature Metabolism, 2022, 4(11): 1551-1559.
Zhang H#, Li Z#, Zhou S, Li S, Ran H, Song Z, Yu T & Yin W*. A fungal NRPS-PKS enzyme catalyses the formation of the flavonoid naringenin. Nature Communications, 2022,13(1):6361.
Zheng T#, Zhang M#, Wu L#, Guo S, Liu X, Zhao J, Xue W, Li J, Liu C, Li X, Jiang Q, Bao J, Zeng J*, Yu T* and Xia C*. Upcycling CO2 into energy-rich long-chain compounds via electrochemical and metabolic engineering. Nature Catalysis. 2022,5:388–396.
Guo S, Wu L, Liu X, Wang B, Yu T*. Developing C1-based metabolic network in methylotrophy for biotransformation. Synthetic Biology Journal, 2022, 3(1):116-137.
Yu T#, Dabirian Y#, Liu Q, Siewers V, Nielsen J*. Strategies and Challenges for Metabolic Rewiring.Current Opinion in Systems Biology, 2019, 15:30-38.
Liu Q., Yu T., Li X., Chen Y., Campbell K., Nielsen J., Chen Y*. Rewiring carbon metabolism in yeast for high level production of aromatic chemicals.Nature Communications, 2019, 10(1):4976.
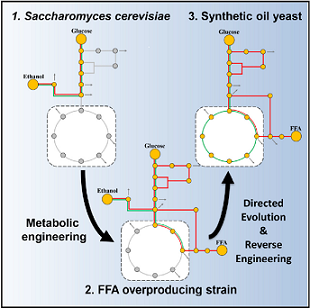
Yu T#, Zhou Y.J.J#, Huang M, Liu Q, Pereira R, David F, Nielsen J*. Reprogramming Yeast Metabolism from Alcoholic Fermentation to Lipogenesis.Cell, 2018, 174(6):1549-1558.
Yu T., Zhou Y.J.J., Wenning L., Liu Q.L., Krivoruchko A., Siewers V., Nielsen, J., and David, F*. Metabolic engineering of Saccharomyces cerevisiae for production of very long chain fatty acid-derived chemicals.Nature Communications, 2017,8:15587.
Yu T., Tao Y., Yang M., Chen P., Gao X., Zhang Y., Zhang T., Chen Z., Hou J., Zhang Y., Ruan K., Wang H., Hu R.G.* Profiling human protein degradome delineates cellular responses to proteasomal inhibition and reveals a feedback mechanism in regulating proteasome homeostasis.Cell Research. 2014, 24(10):1214-1230.
